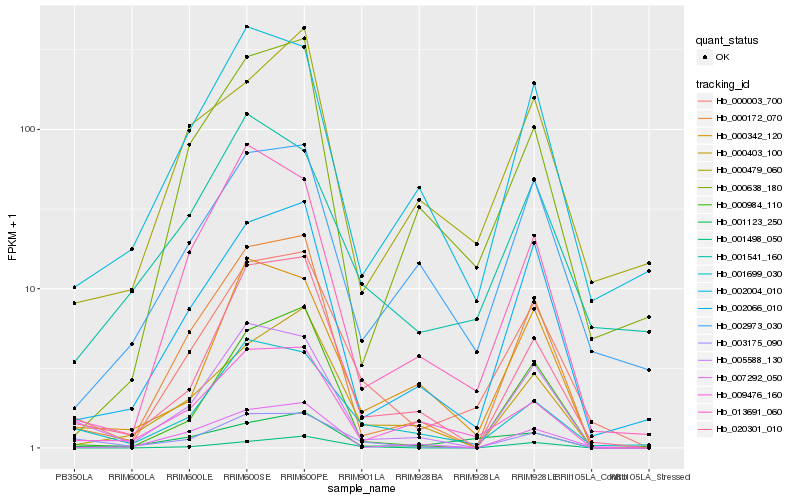
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_700 |
0.0 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 2 |
Hb_002066_010 |
0.1585389264 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001699_030 |
0.1602211768 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_020301_010 |
0.1677796244 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000172_070 |
0.1695810169 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_005588_130 |
0.178304122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002973_030 |
0.1806514415 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 8 |
Hb_002004_010 |
0.184002842 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 9 |
Hb_009476_160 |
0.184493677 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 10 |
Hb_007292_050 |
0.1848066223 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000403_100 |
0.1852161649 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 12 |
Hb_001541_160 |
0.1876792292 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 13 |
Hb_000984_110 |
0.1883490749 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 14 |
Hb_013691_060 |
0.1888731028 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 15 |
Hb_000638_180 |
0.1899411228 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 16 |
Hb_001498_050 |
0.191045337 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 17 |
Hb_000342_120 |
0.1917722439 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 18 |
Hb_003175_090 |
0.1937197027 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 19 |
Hb_001123_250 |
0.1945070653 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 20 |
Hb_000479_060 |
0.1957580711 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |