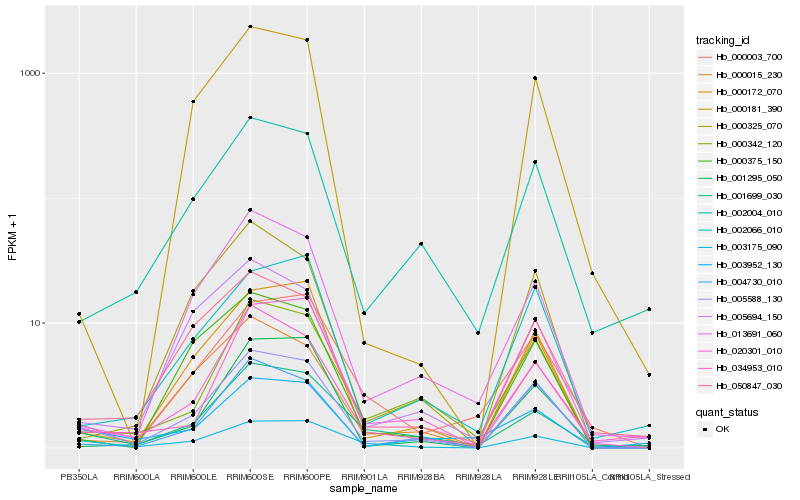
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000172_070 |
0.13408634 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_004730_010 |
0.134156949 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 4 |
Hb_003175_090 |
0.1460454467 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 5 |
Hb_000342_120 |
0.148912962 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 6 |
Hb_020301_010 |
0.1495952931 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_001699_030 |
0.1554439369 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_050847_030 |
0.1568008002 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 9 |
Hb_002004_010 |
0.1604163352 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 10 |
Hb_002066_010 |
0.1614999264 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000015_230 |
0.1627055267 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 12 |
Hb_013691_060 |
0.1655226392 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 13 |
Hb_000375_150 |
0.1667556442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000325_070 |
0.1694137902 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_000181_390 |
0.1738232236 |
- |
- |
unknown [Lotus japonicus] |
| 16 |
Hb_034953_010 |
0.1742559887 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 17 |
Hb_003952_130 |
0.1743966529 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 18 |
Hb_005694_150 |
0.1746525731 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 19 |
Hb_001295_050 |
0.1752062328 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 20 |
Hb_000003_700 |
0.178304122 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |