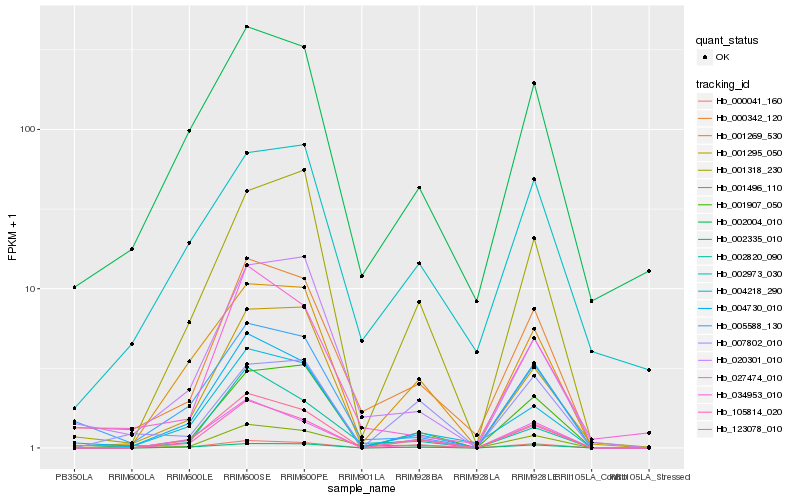
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001496_110 |
0.0 |
- |
- |
hypothetical protein VITISV_044023 [Vitis vinifera] |
| 2 |
Hb_000342_120 |
0.1234497778 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 3 |
Hb_004730_010 |
0.1829897311 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 4 |
Hb_000041_160 |
0.1904703979 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_034953_010 |
0.1923700053 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 6 |
Hb_002004_010 |
0.1951224852 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 7 |
Hb_123078_010 |
0.1974865669 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 8 |
Hb_105814_020 |
0.1988095808 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 9 |
Hb_007802_010 |
0.2002599912 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 10 |
Hb_020301_010 |
0.201695539 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_001907_050 |
0.2041273367 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 12 |
Hb_002973_030 |
0.2083187947 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_005588_130 |
0.2112390762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001269_530 |
0.2148620235 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_001295_050 |
0.2148793227 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 16 |
Hb_002820_090 |
0.2153567926 |
- |
- |
PREDICTED: WAT1-related protein At1g21890-like [Jatropha curcas] |
| 17 |
Hb_004218_290 |
0.2157207713 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 18 |
Hb_027474_010 |
0.2164798003 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 19 |
Hb_002335_010 |
0.2185476599 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 20 |
Hb_001318_230 |
0.2213179375 |
- |
- |
Boron transporter, putative [Ricinus communis] |