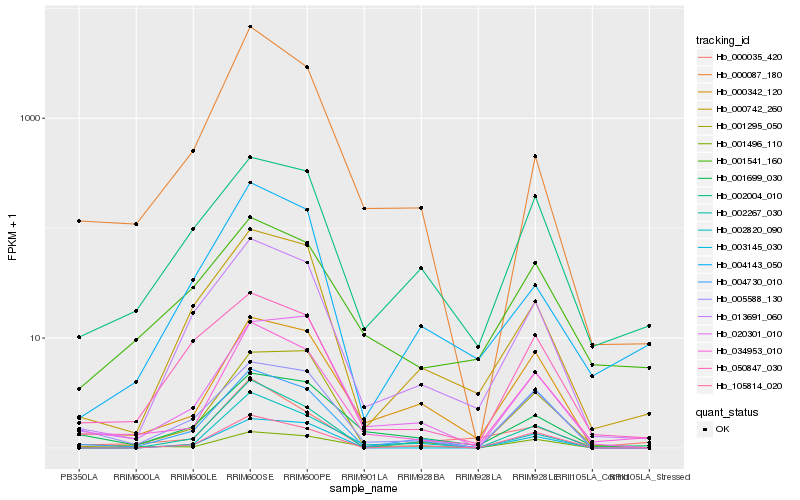
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034953_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 2 |
Hb_000342_120 |
0.1577567366 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 3 |
Hb_005588_130 |
0.1742559887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004730_010 |
0.1757733407 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 5 |
Hb_002004_010 |
0.176099293 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 6 |
Hb_001699_030 |
0.1767585254 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_013691_060 |
0.1811034453 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 8 |
Hb_001295_050 |
0.1812703028 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 9 |
Hb_002820_090 |
0.1830173773 |
- |
- |
PREDICTED: WAT1-related protein At1g21890-like [Jatropha curcas] |
| 10 |
Hb_020301_010 |
0.1859196041 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000035_420 |
0.186192358 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 12 |
Hb_001541_160 |
0.1864036516 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 13 |
Hb_105814_020 |
0.1864364866 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 14 |
Hb_000742_260 |
0.1920428761 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 15 |
Hb_001496_110 |
0.1923700053 |
- |
- |
hypothetical protein VITISV_044023 [Vitis vinifera] |
| 16 |
Hb_000087_180 |
0.1959891073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004143_050 |
0.1975376008 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 18 |
Hb_002267_030 |
0.1977565394 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 19 |
Hb_003145_030 |
0.20062375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_050847_030 |
0.2031118458 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |