| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
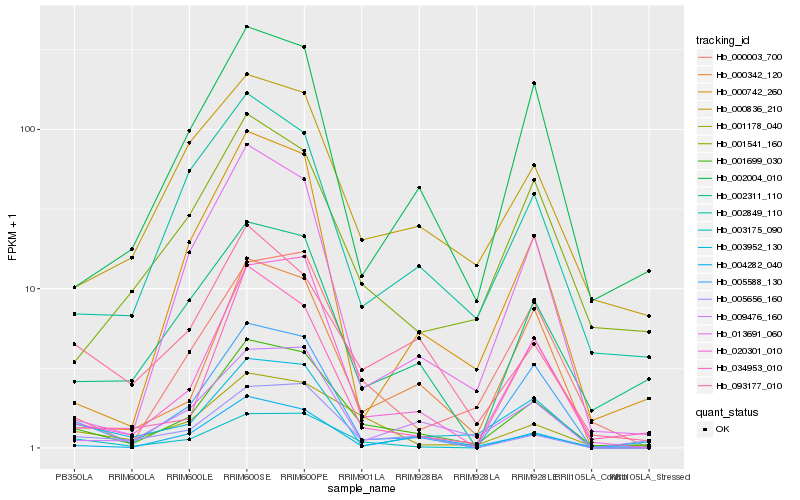
Hb_001699_030 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003175_090 |
0.1417805784 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 3 |
Hb_020301_010 |
0.1486356547 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001178_040 |
0.1503011797 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 5 |
Hb_002311_110 |
0.1533705449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005588_130 |
0.1554439369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004282_040 |
0.1563133986 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_002004_010 |
0.1598531139 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 9 |
Hb_000003_700 |
0.1602211768 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 10 |
Hb_002849_110 |
0.1608972027 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 11 |
Hb_009476_160 |
0.1626464586 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 12 |
Hb_000836_210 |
0.1628596264 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 13 |
Hb_000342_120 |
0.1659827881 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 14 |
Hb_013691_060 |
0.1719773247 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 15 |
Hb_001541_160 |
0.1734036361 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 16 |
Hb_000742_260 |
0.1742486292 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 17 |
Hb_005656_160 |
0.1747860161 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 18 |
Hb_034953_010 |
0.1767585254 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 19 |
Hb_003952_130 |
0.1768375446 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 20 |
Hb_093177_010 |
0.1784004367 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |