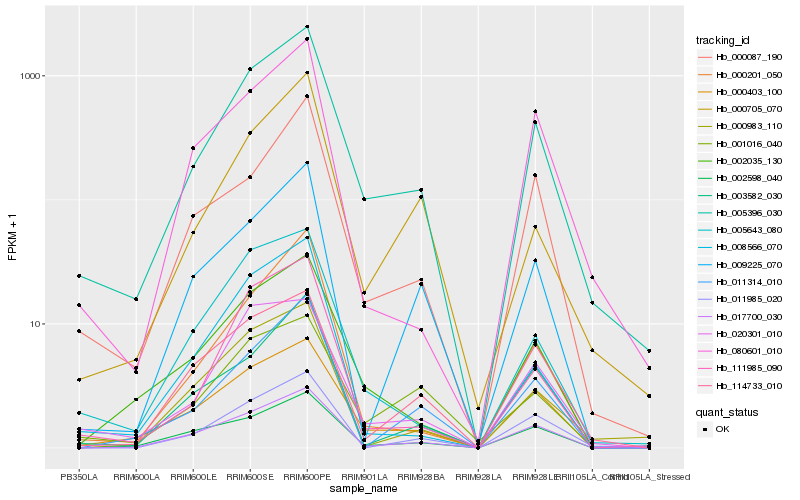
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005396_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000403_100 |
0.1173907136 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 3 |
Hb_017700_030 |
0.1179531272 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 4 |
Hb_002035_130 |
0.1225103199 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 5 |
Hb_111985_090 |
0.1311703921 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 6 |
Hb_005643_080 |
0.1321694898 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 7 |
Hb_020301_010 |
0.1337551601 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_011985_020 |
0.144400346 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002598_040 |
0.1463099281 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 10 |
Hb_000087_190 |
0.146392407 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 11 |
Hb_003582_030 |
0.1479766052 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 12 |
Hb_000705_070 |
0.1482021979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001016_040 |
0.1482360698 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000983_110 |
0.1483296894 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 15 |
Hb_000201_050 |
0.1496644072 |
- |
- |
- |
| 16 |
Hb_011314_010 |
0.1506035534 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_009225_070 |
0.1510389254 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 18 |
Hb_114733_010 |
0.153529835 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 19 |
Hb_080601_010 |
0.1536780061 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 20 |
Hb_008566_070 |
0.1539358704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |