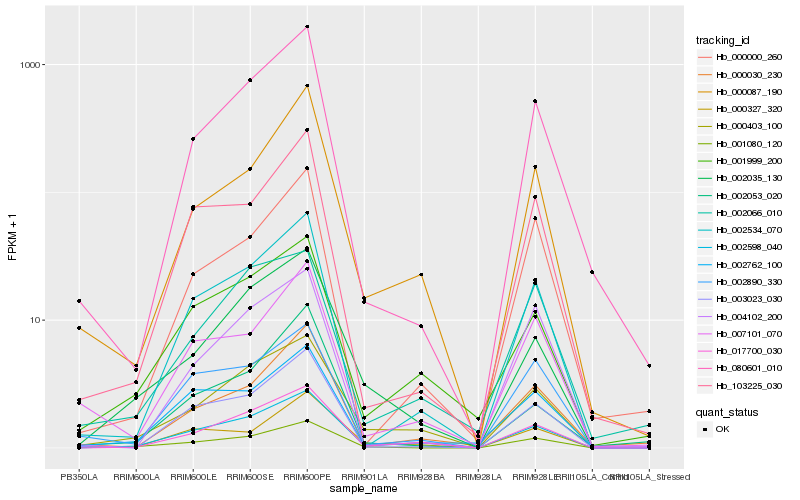
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_120 |
0.0 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 2 |
Hb_080601_010 |
0.1520004163 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 3 |
Hb_103225_030 |
0.1583066046 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_002035_130 |
0.1595605958 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 5 |
Hb_000030_230 |
0.1645528477 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 6 |
Hb_000403_100 |
0.1654219323 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 7 |
Hb_003023_030 |
0.1674874655 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_007101_070 |
0.1675516717 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 9 |
Hb_002534_070 |
0.1678387431 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 10 |
Hb_002598_040 |
0.1683246646 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 11 |
Hb_002762_100 |
0.1689316007 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000327_320 |
0.1691790957 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g39360 [Jatropha curcas] |
| 13 |
Hb_000000_260 |
0.1699825805 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 14 |
Hb_002890_330 |
0.170670466 |
- |
- |
caspase, putative [Ricinus communis] |
| 15 |
Hb_001999_200 |
0.1741346398 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000087_190 |
0.1742746686 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 17 |
Hb_002053_020 |
0.1767597146 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 18 |
Hb_002066_010 |
0.1797087831 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_017700_030 |
0.183729505 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 20 |
Hb_004102_200 |
0.1839380531 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |