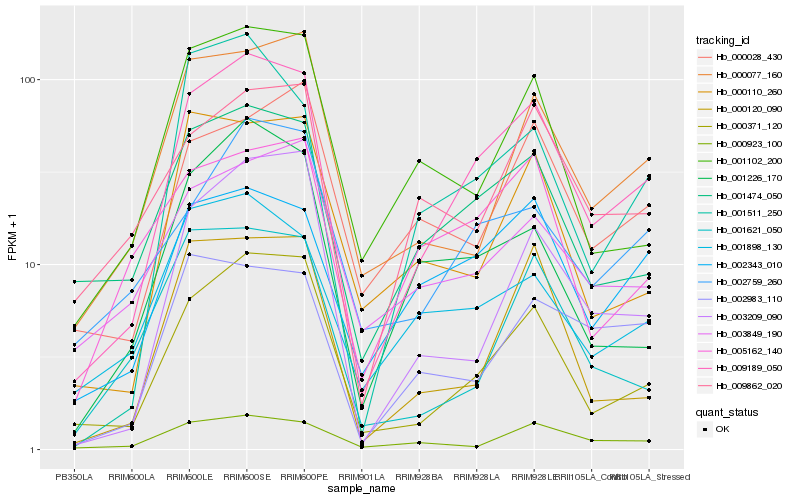
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009189_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648080 [Jatropha curcas] |
| 2 |
Hb_000371_120 |
0.1169139399 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_001474_050 |
0.1430489316 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 4 |
Hb_002343_010 |
0.1496315205 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000077_160 |
0.1627802807 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 6 |
Hb_002759_260 |
0.1648005527 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 7 |
Hb_003209_090 |
0.1659989604 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 8 |
Hb_001511_250 |
0.1660731736 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 9 |
Hb_001102_200 |
0.1684400255 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 10 |
Hb_009862_020 |
0.1687192284 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 11 |
Hb_000923_100 |
0.1703314768 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 12 |
Hb_000120_090 |
0.1711051255 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 13 |
Hb_001226_170 |
0.1721088682 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 14 |
Hb_001898_130 |
0.1723009575 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_002983_110 |
0.1739089886 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 16 |
Hb_000028_430 |
0.1744098591 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005162_140 |
0.1746385465 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 18 |
Hb_000110_260 |
0.176814752 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 19 |
Hb_003849_190 |
0.1774540238 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 20 |
Hb_001621_050 |
0.1849320396 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |