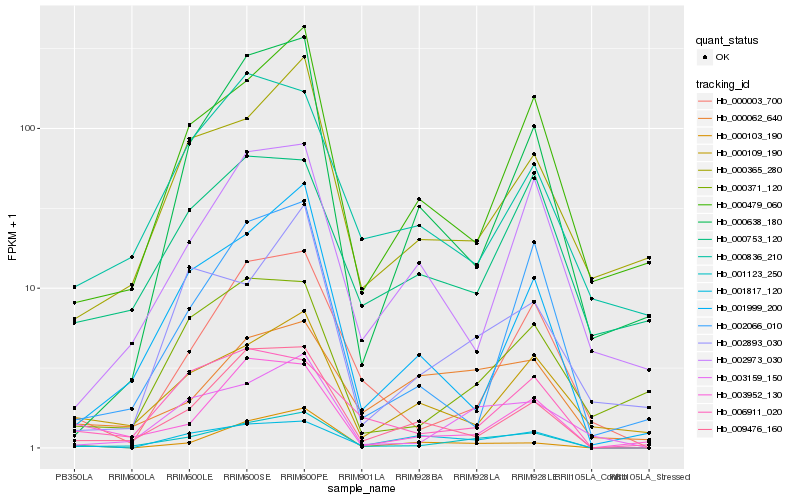
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_250 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 2 |
Hb_003159_150 |
0.14448225 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 3 |
Hb_009476_160 |
0.1732507938 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 4 |
Hb_000479_060 |
0.1869630788 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 5 |
Hb_001999_200 |
0.1927278801 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000003_700 |
0.1945070653 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_000365_280 |
0.1979517229 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 8 |
Hb_003952_130 |
0.2090438056 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 9 |
Hb_000638_180 |
0.2099501328 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 10 |
Hb_000109_190 |
0.2106917936 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 11 |
Hb_002893_030 |
0.2110389102 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 12 |
Hb_000371_120 |
0.212380452 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001817_120 |
0.2131508731 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_002066_010 |
0.2142341174 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006911_020 |
0.2148535352 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_002973_030 |
0.2187133819 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000062_640 |
0.218979485 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000103_190 |
0.2217057559 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 19 |
Hb_000836_210 |
0.2227642219 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 20 |
Hb_000753_120 |
0.223034113 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |