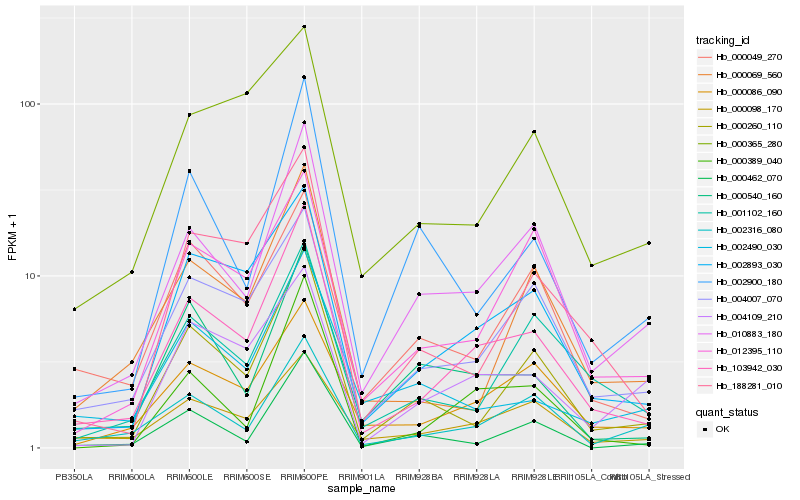
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103942_030 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_002893_030 |
0.1250955578 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 3 |
Hb_010883_180 |
0.1318055871 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 4 |
Hb_000260_110 |
0.1404357752 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_000389_040 |
0.1462442719 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g03250-like [Jatropha curcas] |
| 6 |
Hb_004007_070 |
0.1499273108 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 7 |
Hb_000086_090 |
0.153324604 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_000098_170 |
0.1537289006 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_188281_010 |
0.1620802213 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000069_560 |
0.1668773739 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 11 |
Hb_000540_160 |
0.1715932574 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 12 |
Hb_001102_160 |
0.1753096309 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 13 |
Hb_012395_110 |
0.1758534399 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 14 |
Hb_002316_080 |
0.1773858389 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 15 |
Hb_000049_270 |
0.1786778165 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002900_180 |
0.182714843 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 17 |
Hb_000365_280 |
0.1864953881 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 18 |
Hb_002490_030 |
0.1904058913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004109_210 |
0.1923381989 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000462_070 |
0.1947744489 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |