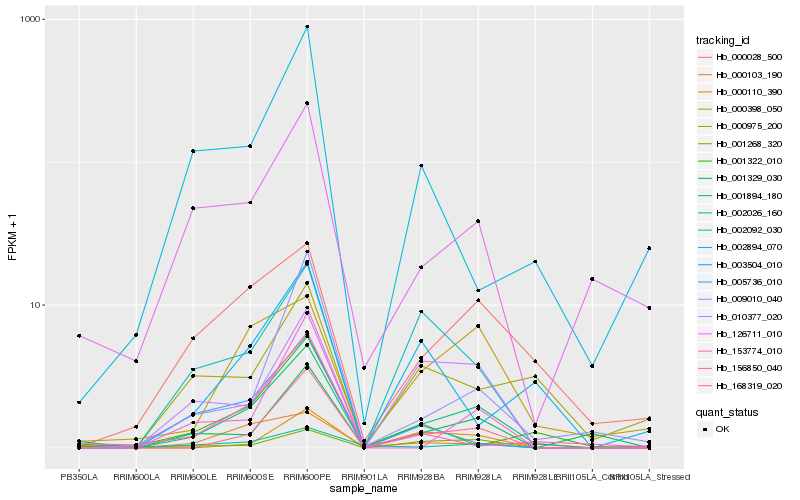
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_180 |
0.0 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 2 |
Hb_168319_020 |
0.1956702191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000398_050 |
0.2210961577 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 4 |
Hb_000028_500 |
0.227795773 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_126711_010 |
0.2316584592 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 6 |
Hb_010377_020 |
0.2316971404 |
- |
- |
- |
| 7 |
Hb_001329_030 |
0.2329425521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002026_160 |
0.239987552 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 9 |
Hb_001268_320 |
0.2483226178 |
- |
- |
PREDICTED: uncharacterized protein LOC104887816 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_002092_030 |
0.2512129652 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 11 |
Hb_009010_040 |
0.2533333007 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000110_390 |
0.2602902597 |
- |
- |
PREDICTED: uncharacterized protein LOC103338992 [Prunus mume] |
| 13 |
Hb_000975_200 |
0.2615744749 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000103_190 |
0.263450828 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 15 |
Hb_153774_010 |
0.2657099388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001322_010 |
0.2712728786 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 17 |
Hb_003504_010 |
0.2725120406 |
- |
- |
hypothetical protein POPTR_0013s04290g [Populus trichocarpa] |
| 18 |
Hb_156850_040 |
0.2729891364 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 19 |
Hb_002894_070 |
0.2738776885 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 20 |
Hb_005736_010 |
0.2754967596 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |