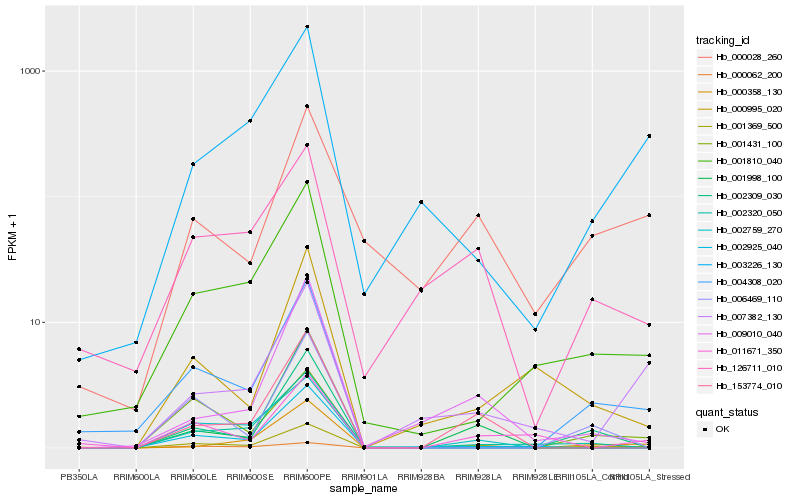
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_500 |
0.0 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 2 |
Hb_004308_020 |
0.2031315071 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 3 |
Hb_001810_040 |
0.2133881229 |
- |
- |
- |
| 4 |
Hb_002320_050 |
0.2136406486 |
- |
- |
clathrin assembly family protein [Populus trichocarpa] |
| 5 |
Hb_153774_010 |
0.2148613795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_126711_010 |
0.2155684266 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 7 |
Hb_000358_130 |
0.2170269161 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 8 |
Hb_002759_270 |
0.2197762606 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_009010_040 |
0.2313429641 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 10 |
Hb_002309_030 |
0.2352586139 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 11 |
Hb_006469_110 |
0.2367646843 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 12 |
Hb_001998_100 |
0.2394541178 |
- |
- |
- |
| 13 |
Hb_001431_100 |
0.2440101202 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 [Jatropha curcas] |
| 14 |
Hb_000995_020 |
0.2442932415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003226_130 |
0.2454937542 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 16 |
Hb_000028_260 |
0.2462791032 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 17 |
Hb_002925_040 |
0.2518819665 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 18 |
Hb_011671_350 |
0.2546680553 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 19 |
Hb_000062_200 |
0.2547204696 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_007382_130 |
0.2570262024 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |