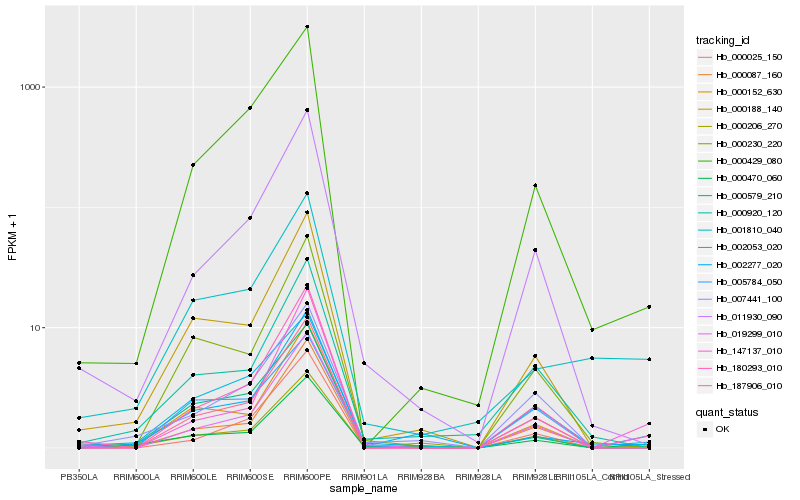
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000470_060 |
0.0 |
- |
- |
PREDICTED: abietadienol/abietadienal oxidase [Jatropha curcas] |
| 2 |
Hb_000188_140 |
0.1349447308 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 3 |
Hb_180293_010 |
0.1354776047 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 4 |
Hb_002053_020 |
0.1431044142 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 5 |
Hb_000152_630 |
0.1439099109 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 6 |
Hb_000206_270 |
0.1440405082 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000230_220 |
0.1464465059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000087_160 |
0.1482963503 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_187906_010 |
0.1504594421 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 10 |
Hb_019299_010 |
0.1523496753 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000429_080 |
0.1537034974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011930_090 |
0.1545410283 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 13 |
Hb_001810_040 |
0.155784414 |
- |
- |
- |
| 14 |
Hb_005784_050 |
0.1572032405 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_147137_010 |
0.1574754293 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 16 |
Hb_000579_210 |
0.15796043 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 17 |
Hb_002277_020 |
0.1581334226 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 18 |
Hb_000920_120 |
0.1592910582 |
- |
- |
- |
| 19 |
Hb_000025_150 |
0.1598973082 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 20 |
Hb_007441_100 |
0.1599792296 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |