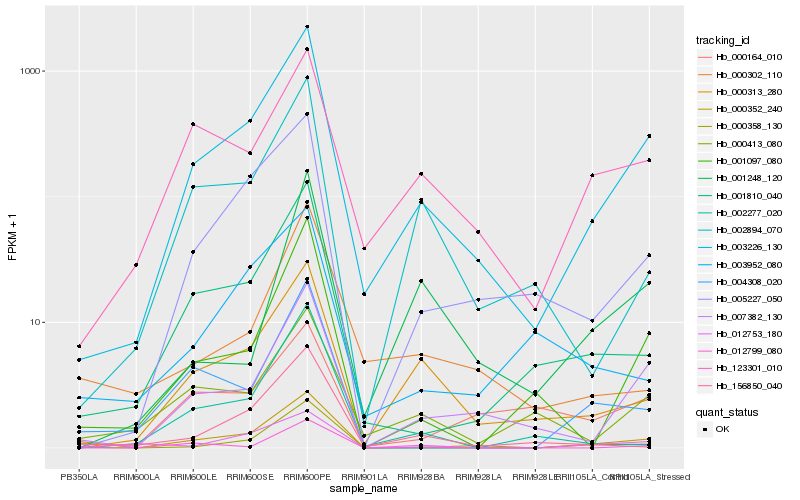
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_130 |
0.0 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 2 |
Hb_007382_130 |
0.1564517517 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_005227_050 |
0.1612082317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000313_280 |
0.1725238749 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 5 |
Hb_001810_040 |
0.1903631478 |
- |
- |
- |
| 6 |
Hb_123301_010 |
0.1943606913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001097_080 |
0.1972839498 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 8 |
Hb_000358_130 |
0.1975427418 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 9 |
Hb_000413_080 |
0.2029710985 |
- |
- |
- |
| 10 |
Hb_002894_070 |
0.2047227833 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 11 |
Hb_000352_240 |
0.2106431008 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_012753_180 |
0.2143709576 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 13 |
Hb_156850_040 |
0.2318952264 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 14 |
Hb_002277_020 |
0.2338454887 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 15 |
Hb_004308_020 |
0.2380581495 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 16 |
Hb_003952_080 |
0.2390409704 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000302_110 |
0.2393888009 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001248_120 |
0.2402971429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000164_010 |
0.2405795571 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 20 |
Hb_012799_080 |
0.2433351207 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |