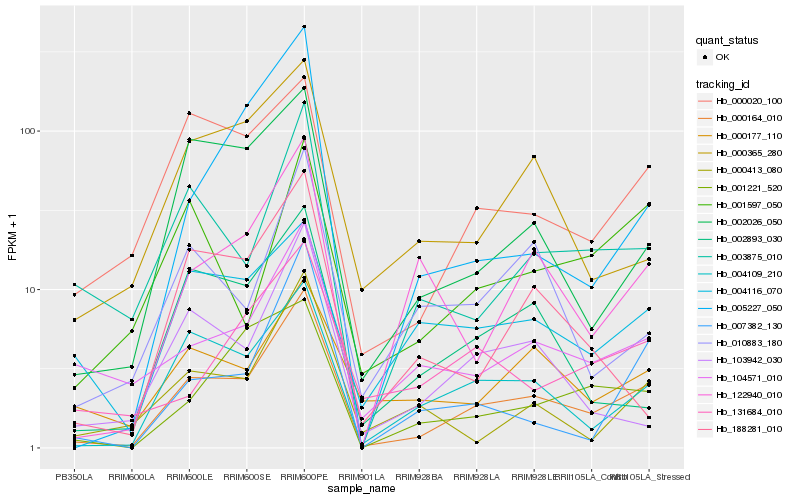
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000164_010 |
0.0 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 2 |
Hb_004109_210 |
0.1609295701 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_003875_010 |
0.1970360021 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 4 |
Hb_104571_010 |
0.1987044117 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 5 |
Hb_002026_050 |
0.199799019 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 6 |
Hb_000020_100 |
0.2032646049 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 7 |
Hb_122940_010 |
0.2038464377 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_103942_030 |
0.2078945175 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_007382_130 |
0.2140828864 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 10 |
Hb_000177_110 |
0.2169468034 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 11 |
Hb_010883_180 |
0.2172141462 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 12 |
Hb_002893_030 |
0.2174071488 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 13 |
Hb_000413_080 |
0.2181040517 |
- |
- |
- |
| 14 |
Hb_000365_280 |
0.2209702574 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 15 |
Hb_001221_520 |
0.2223750391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005227_050 |
0.2232541904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004116_070 |
0.2246374685 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 18 |
Hb_188281_010 |
0.2255175178 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 19 |
Hb_001597_050 |
0.2263666174 |
- |
- |
- |
| 20 |
Hb_131684_010 |
0.2278438226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |