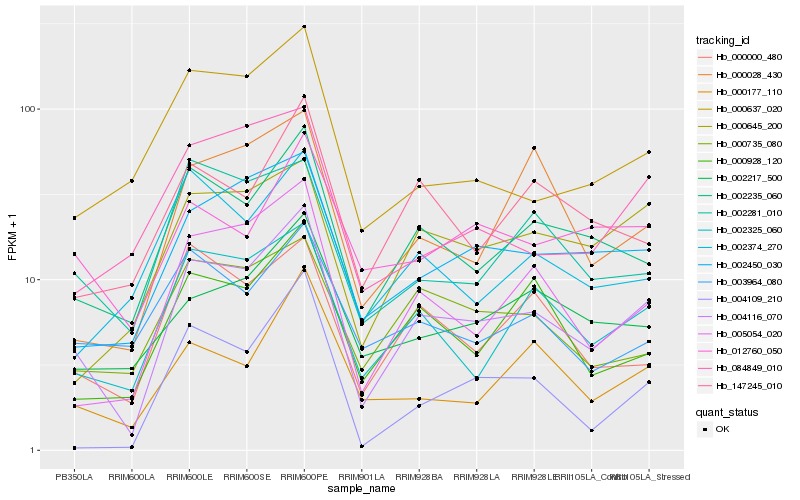
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 2 |
Hb_002235_060 |
0.1331055787 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_005054_020 |
0.1369128587 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 4 |
Hb_002325_060 |
0.1429913907 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 5 |
Hb_002450_030 |
0.1493892711 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 6 |
Hb_000928_120 |
0.1639256815 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 7 |
Hb_000645_200 |
0.167649532 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 8 |
Hb_000000_480 |
0.1687245749 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002217_500 |
0.1687457048 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 10 |
Hb_000177_110 |
0.1704990714 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 11 |
Hb_000735_080 |
0.1732602683 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 12 |
Hb_000637_020 |
0.1741503206 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 13 |
Hb_147245_010 |
0.1743019869 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 14 |
Hb_002281_010 |
0.1778170815 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 15 |
Hb_004109_210 |
0.1792396779 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002374_270 |
0.1794770933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012760_050 |
0.1811322674 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 18 |
Hb_084849_010 |
0.181266624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000028_430 |
0.1820686106 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003964_080 |
0.1821454769 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |