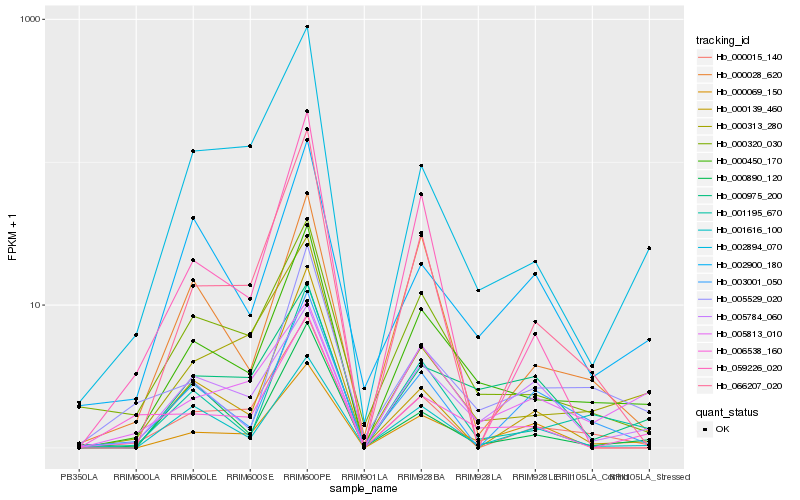
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000450_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001195_670 |
0.128088112 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 3 |
Hb_000320_030 |
0.1513358458 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 4 |
Hb_000313_280 |
0.1601924882 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 5 |
Hb_000015_140 |
0.1674037064 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 6 |
Hb_005529_020 |
0.1779540559 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 7 |
Hb_002894_070 |
0.1785569753 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 8 |
Hb_066207_020 |
0.1849636289 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 9 |
Hb_000975_200 |
0.1868184481 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001616_100 |
0.190086365 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 11 |
Hb_002900_180 |
0.1908018505 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 12 |
Hb_006538_160 |
0.1911234824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005784_060 |
0.1929317269 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 14 |
Hb_000028_620 |
0.196577635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_059226_020 |
0.2010522241 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 16 |
Hb_000890_120 |
0.203888557 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003001_050 |
0.2047094699 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 18 |
Hb_000139_460 |
0.20745232 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 19 |
Hb_005813_010 |
0.2075368884 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000069_150 |
0.2098184873 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |