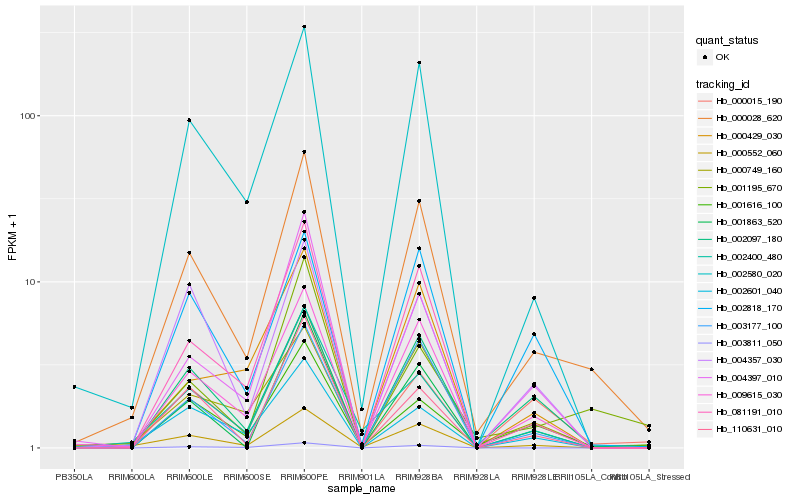
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_620 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009615_030 |
0.1233472948 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000552_060 |
0.1345566095 |
- |
- |
PREDICTED: theobromine synthase 2-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_002580_020 |
0.1357047755 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 5 |
Hb_081191_010 |
0.1452925038 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 6 |
Hb_000749_160 |
0.1505559726 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_002097_180 |
0.1534265664 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003177_100 |
0.1589493432 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 9 |
Hb_001616_100 |
0.1594113127 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 10 |
Hb_002601_040 |
0.1635468103 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 11 |
Hb_002400_480 |
0.1688964918 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 12 |
Hb_000015_190 |
0.1701824882 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 13 |
Hb_001863_520 |
0.1705955014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004397_010 |
0.1761714412 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002818_170 |
0.1764168846 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 16 |
Hb_001195_670 |
0.1775412763 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 17 |
Hb_004357_030 |
0.1786448733 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_003811_050 |
0.1796784849 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 19 |
Hb_000429_030 |
0.1803197626 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 20 |
Hb_110631_010 |
0.1823492721 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |