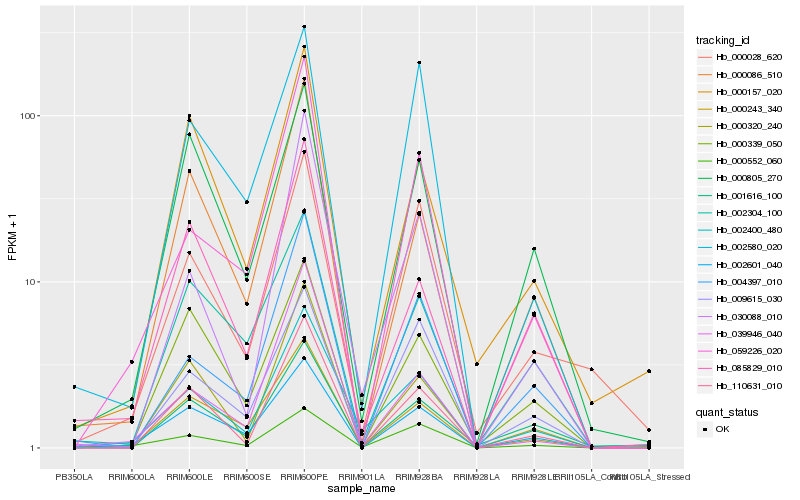
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_480 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 2 |
Hb_002601_040 |
0.1440534387 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |
| 3 |
Hb_000243_340 |
0.1499466445 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 4 |
Hb_000552_060 |
0.1552109348 |
- |
- |
PREDICTED: theobromine synthase 2-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_110631_010 |
0.1568916088 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 6 |
Hb_030088_010 |
0.1577908264 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 7 |
Hb_004397_010 |
0.1618931962 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_059226_020 |
0.1637776935 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 9 |
Hb_000086_510 |
0.1653725338 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_009615_030 |
0.167849443 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000028_620 |
0.1688964918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001616_100 |
0.1696796325 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 13 |
Hb_000320_240 |
0.1716398582 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 14 |
Hb_002304_100 |
0.1745749275 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 15 |
Hb_000339_050 |
0.1761253981 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 16 |
Hb_000157_020 |
0.1764350773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_085829_010 |
0.176555859 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 18 |
Hb_039946_040 |
0.1774029542 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 19 |
Hb_000805_270 |
0.1778173721 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 20 |
Hb_002580_020 |
0.1823270861 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |