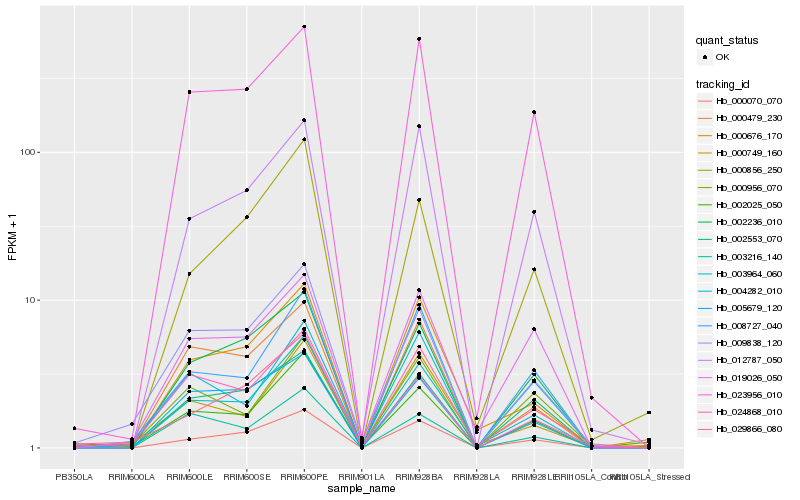
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_060 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 2 |
Hb_000070_070 |
0.100386744 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 3 |
Hb_008727_040 |
0.1118118547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002025_050 |
0.1136825822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_023956_010 |
0.1144955453 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 6 |
Hb_000676_170 |
0.1150028803 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 7 |
Hb_000749_160 |
0.1202575292 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_002236_010 |
0.1204121675 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 9 |
Hb_012787_050 |
0.1211888917 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 10 |
Hb_000479_230 |
0.1242200973 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 11 |
Hb_009838_120 |
0.1276110225 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 12 |
Hb_029866_080 |
0.1311612317 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 13 |
Hb_000856_250 |
0.1318970373 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_002553_070 |
0.1362717805 |
- |
- |
PREDICTED: RING-H2 finger protein ATL16 [Jatropha curcas] |
| 15 |
Hb_005679_120 |
0.1362924496 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 16 |
Hb_000956_070 |
0.1378302902 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 17 |
Hb_019026_050 |
0.1379909439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004282_010 |
0.1394074943 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 19 |
Hb_003216_140 |
0.1401940616 |
- |
- |
F-box protein SNE, putative [Ricinus communis] |
| 20 |
Hb_024868_010 |
0.1423898276 |
- |
- |
protein kinase, putative [Ricinus communis] |