| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
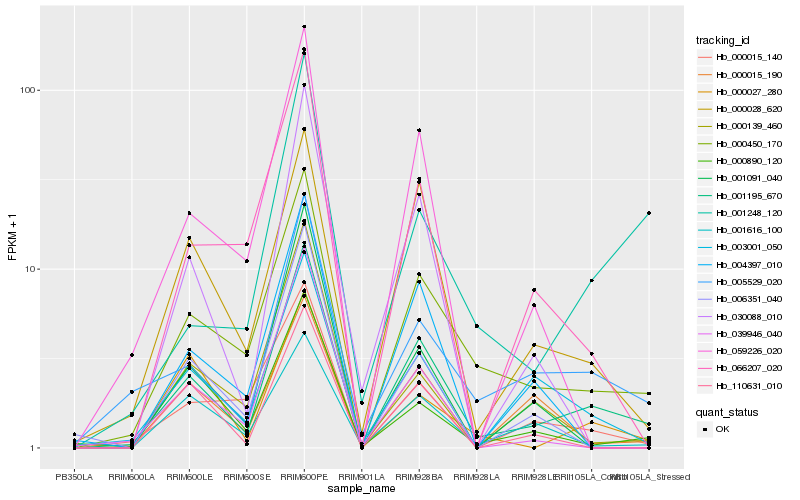
Hb_001195_670 |
0.0 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 2 |
Hb_000450_170 |
0.128088112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005529_020 |
0.1577648504 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 4 |
Hb_000028_620 |
0.1775412763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003001_050 |
0.1807198971 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 6 |
Hb_000015_140 |
0.1882195183 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 7 |
Hb_066207_020 |
0.1905997967 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 8 |
Hb_059226_020 |
0.1952407944 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 9 |
Hb_030088_010 |
0.1961637664 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 10 |
Hb_004397_010 |
0.2015224555 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000139_460 |
0.2039143862 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 12 |
Hb_039946_040 |
0.2044573716 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 13 |
Hb_001616_100 |
0.2065091784 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 14 |
Hb_001091_040 |
0.2087602171 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 15 |
Hb_000027_280 |
0.2103568085 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 16 |
Hb_110631_010 |
0.2108605087 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 17 |
Hb_006351_040 |
0.2110817804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000890_120 |
0.2119895785 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001248_120 |
0.2130892592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000015_190 |
0.213415334 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |