| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
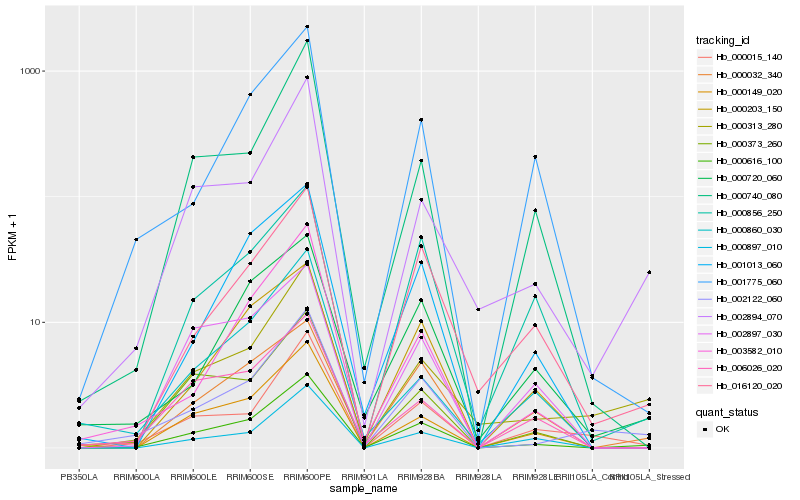
Hb_000616_100 |
0.0 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000149_020 |
0.126640403 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 3 |
Hb_002122_060 |
0.1299344875 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_002894_070 |
0.1341897767 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 5 |
Hb_000373_260 |
0.1390365407 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 6 |
Hb_000740_080 |
0.145180237 |
- |
- |
- |
| 7 |
Hb_003582_010 |
0.1472085414 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 8 |
Hb_001013_060 |
0.1492475711 |
- |
- |
- |
| 9 |
Hb_016120_020 |
0.1517155064 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 10 |
Hb_000032_340 |
0.1524645567 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 11 |
Hb_000313_280 |
0.1525673193 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 12 |
Hb_006026_020 |
0.1528553719 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48800 [Jatropha curcas] |
| 13 |
Hb_000203_150 |
0.1548630293 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 14 |
Hb_000897_010 |
0.1577367777 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 15 |
Hb_000720_060 |
0.1604520072 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 16 |
Hb_001775_060 |
0.1629266443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000015_140 |
0.1629606247 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 18 |
Hb_002897_030 |
0.1636566927 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 19 |
Hb_000860_030 |
0.1651855822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000856_250 |
0.1654299135 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |