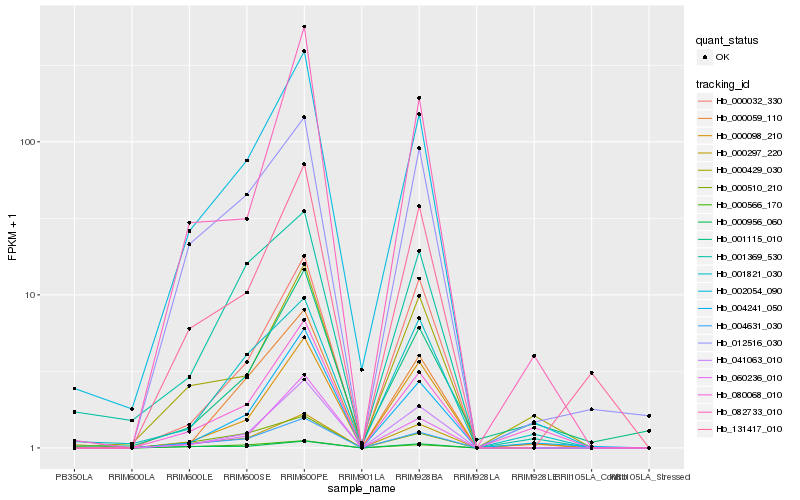
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002054_090 |
0.0 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 2 |
Hb_004631_030 |
0.0906408344 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 3 |
Hb_041063_010 |
0.1078358378 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 4 |
Hb_000297_220 |
0.1130198806 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 5 |
Hb_060236_010 |
0.1214503907 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 6 |
Hb_004241_050 |
0.1325181554 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 7 |
Hb_000059_110 |
0.1345931169 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 8 |
Hb_012516_030 |
0.1353651718 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 9 |
Hb_131417_010 |
0.1362779418 |
- |
- |
hypothetical protein POPTR_0011s066602g, partial [Populus trichocarpa] |
| 10 |
Hb_000510_210 |
0.1418986232 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000956_060 |
0.1424409208 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001369_530 |
0.145528186 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 13 |
Hb_000429_030 |
0.1456068338 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_082733_010 |
0.1468194445 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 15 |
Hb_000566_170 |
0.1468713618 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 16 |
Hb_000032_330 |
0.1494882347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_080068_010 |
0.1506403798 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 18 |
Hb_000098_210 |
0.1557255294 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 19 |
Hb_001821_030 |
0.1557934928 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 20 |
Hb_001115_010 |
0.1568638531 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |