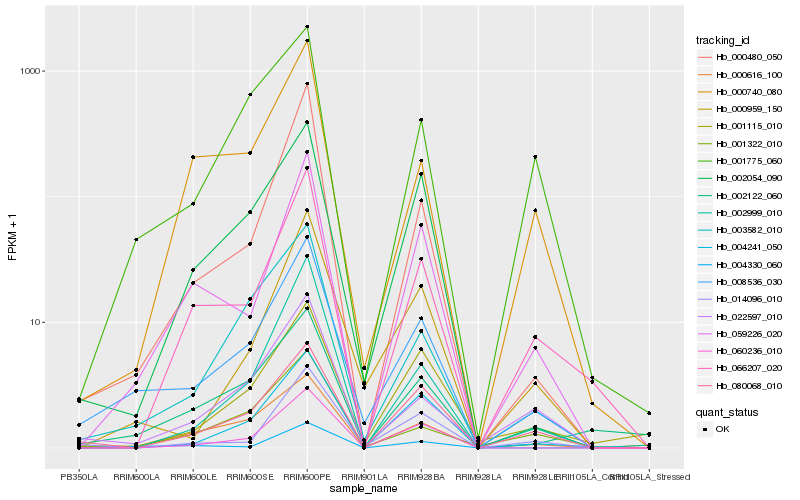
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008536_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 2 |
Hb_003582_010 |
0.1546482463 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 3 |
Hb_000959_150 |
0.1654464592 |
- |
- |
PREDICTED: uncharacterized protein LOC104879927 [Vitis vinifera] |
| 4 |
Hb_059226_020 |
0.1671811592 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 5 |
Hb_022597_010 |
0.1698791167 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 6 |
Hb_080068_010 |
0.1723136138 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 7 |
Hb_002054_090 |
0.1725540965 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 8 |
Hb_000480_050 |
0.1758079341 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 9 |
Hb_002122_060 |
0.1762477494 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000616_100 |
0.1763904772 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001322_010 |
0.1787579362 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 12 |
Hb_004330_060 |
0.1796304294 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 13 |
Hb_014096_010 |
0.1798367273 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001775_060 |
0.1812335051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004241_050 |
0.1838166883 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 16 |
Hb_060236_010 |
0.1840198715 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 17 |
Hb_066207_020 |
0.1845090112 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 18 |
Hb_000740_080 |
0.1864984885 |
- |
- |
- |
| 19 |
Hb_001115_010 |
0.1875228505 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |
| 20 |
Hb_002999_010 |
0.1892733543 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |