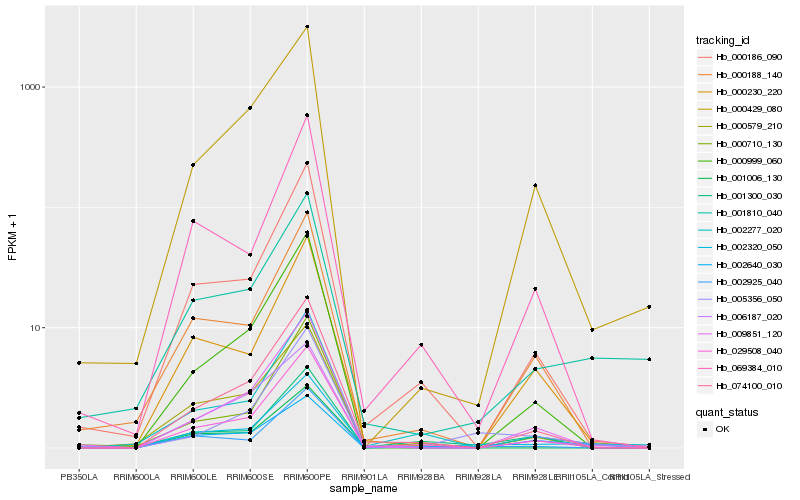
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002320_050 |
0.0 |
- |
- |
clathrin assembly family protein [Populus trichocarpa] |
| 2 |
Hb_029508_040 |
0.1291690313 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_002925_040 |
0.1423787539 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 4 |
Hb_001810_040 |
0.1453397817 |
- |
- |
- |
| 5 |
Hb_000186_090 |
0.1458983142 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 6 |
Hb_005356_050 |
0.1461444953 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 7 |
Hb_074100_010 |
0.1478877768 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 8 |
Hb_002277_020 |
0.1517715405 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 9 |
Hb_000579_210 |
0.1569409481 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 10 |
Hb_002640_030 |
0.1599970278 |
- |
- |
hypothetical protein POPTR_0014s05270g [Populus trichocarpa] |
| 11 |
Hb_000710_130 |
0.1600358722 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_000230_220 |
0.1607869105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006187_020 |
0.1626817241 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |
| 14 |
Hb_069384_010 |
0.163827019 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000429_080 |
0.1658534956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000999_060 |
0.1670146757 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 17 |
Hb_001300_030 |
0.1684689728 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |
| 18 |
Hb_000188_140 |
0.1694897171 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 19 |
Hb_009851_120 |
0.1698245922 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 20 |
Hb_001006_130 |
0.1713906789 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |