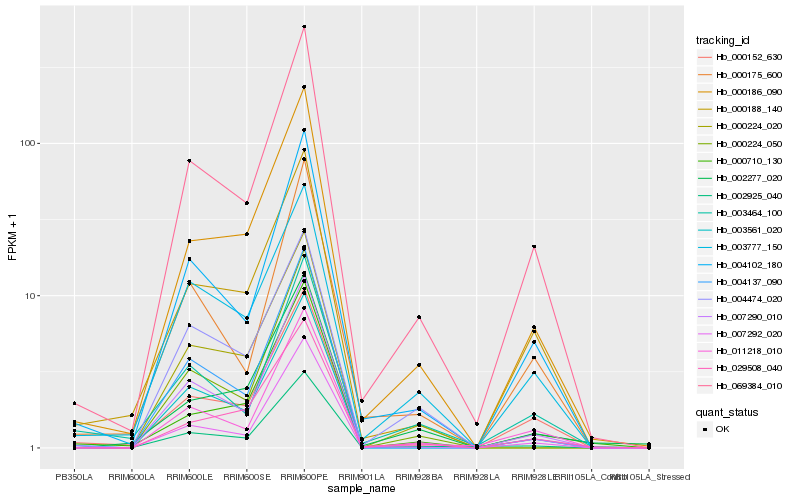
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002925_040 |
0.0 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 2 |
Hb_069384_010 |
0.0849953004 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000186_090 |
0.0929676502 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 4 |
Hb_011218_010 |
0.0940216049 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 5 |
Hb_004102_180 |
0.1000363782 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_007290_010 |
0.1059198843 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_007292_020 |
0.1102234224 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 8 |
Hb_004137_090 |
0.112082621 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 9 |
Hb_003561_020 |
0.1174456582 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 10 |
Hb_000188_140 |
0.1201870498 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 11 |
Hb_000152_630 |
0.1210556143 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 12 |
Hb_000224_050 |
0.1230558111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000175_600 |
0.1277779825 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 14 |
Hb_000710_130 |
0.1281699748 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_029508_040 |
0.1295628947 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 16 |
Hb_003464_100 |
0.1302610187 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 17 |
Hb_000224_020 |
0.1334708853 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 18 |
Hb_003777_150 |
0.1339584931 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004474_020 |
0.1342477127 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 20 |
Hb_002277_020 |
0.134720822 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |