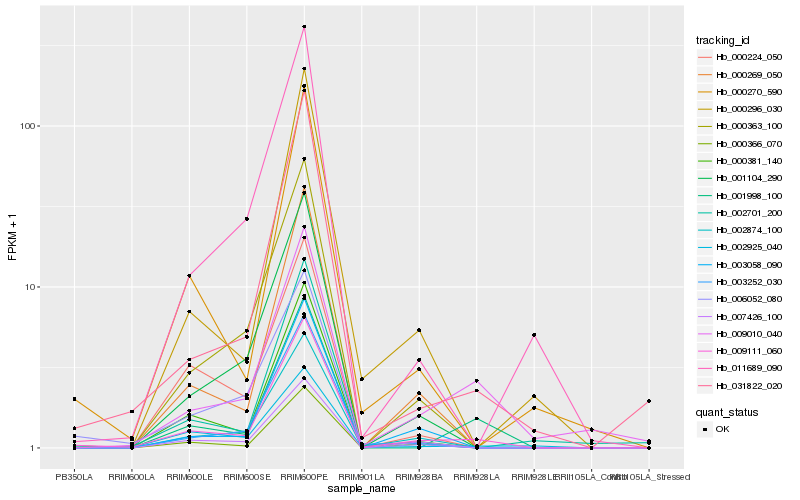
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009111_060 |
0.0 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 2 |
Hb_031822_020 |
0.1312717909 |
- |
- |
PREDICTED: uncharacterized protein LOC104613080 [Nelumbo nucifera] |
| 3 |
Hb_000381_140 |
0.133977972 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 4 |
Hb_002874_100 |
0.1373374094 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000269_050 |
0.1376577159 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 6 |
Hb_000366_070 |
0.1391327663 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.2 [Jatropha curcas] |
| 7 |
Hb_000270_590 |
0.1403201197 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 8 |
Hb_003252_030 |
0.1424844137 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 9 |
Hb_001998_100 |
0.1450506672 |
- |
- |
- |
| 10 |
Hb_002925_040 |
0.1452967451 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 11 |
Hb_000363_100 |
0.1456160752 |
- |
- |
- |
| 12 |
Hb_001104_290 |
0.1462039761 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 13 |
Hb_007426_100 |
0.1485023361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003058_090 |
0.1488481856 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 15 |
Hb_009010_040 |
0.1502857592 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 16 |
Hb_000296_030 |
0.1556772059 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 17 |
Hb_002701_200 |
0.1590545423 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 18 |
Hb_006052_080 |
0.1609280866 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH140-like [Populus euphratica] |
| 19 |
Hb_011689_090 |
0.1611777657 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 20 |
Hb_000224_050 |
0.1612616632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |