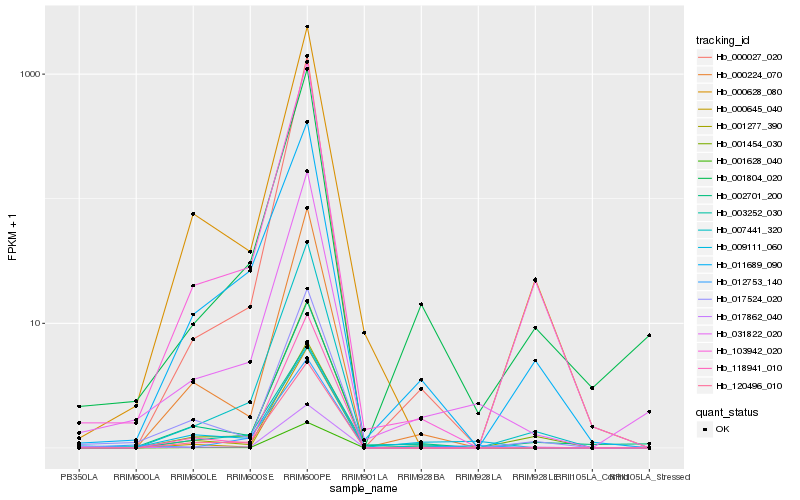
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031822_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104613080 [Nelumbo nucifera] |
| 2 |
Hb_001804_020 |
0.0867759707 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 3 |
Hb_002701_200 |
0.1061913124 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 4 |
Hb_012753_140 |
0.1110663191 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 5 |
Hb_007441_320 |
0.1127896403 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 6 |
Hb_120496_010 |
0.1173682474 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 7 |
Hb_000224_070 |
0.1189031899 |
- |
- |
- |
| 8 |
Hb_103942_020 |
0.1192450718 |
- |
- |
unknown [Zea mays] |
| 9 |
Hb_000628_080 |
0.1231181611 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 10 |
Hb_001277_390 |
0.1236020533 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 11 |
Hb_118941_010 |
0.1243910345 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_017524_020 |
0.1245089016 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 13 |
Hb_000027_020 |
0.1245868526 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 14 |
Hb_001628_040 |
0.1257652768 |
- |
- |
PREDICTED: beta-galactosidase 13-like [Populus euphratica] |
| 15 |
Hb_017862_040 |
0.1261040293 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 16 |
Hb_011689_090 |
0.1296972284 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 17 |
Hb_001454_030 |
0.1306510303 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 18 |
Hb_009111_060 |
0.1312717909 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 19 |
Hb_003252_030 |
0.1317636242 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 20 |
Hb_000645_040 |
0.131781034 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |