| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
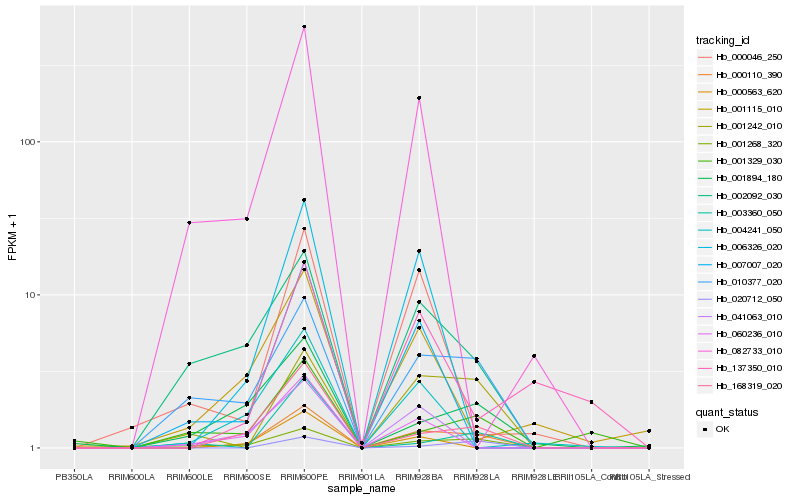
Hb_000110_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103338992 [Prunus mume] |
| 2 |
Hb_168319_020 |
0.1872121683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010377_020 |
0.1939420469 |
- |
- |
- |
| 4 |
Hb_001268_320 |
0.2198209736 |
- |
- |
PREDICTED: uncharacterized protein LOC104887816 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001242_010 |
0.2362430092 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 6 |
Hb_002092_030 |
0.2399684934 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 7 |
Hb_001894_180 |
0.2602902597 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 8 |
Hb_020712_050 |
0.2638677786 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 9 |
Hb_000563_620 |
0.289084693 |
- |
- |
hypothetical protein JCGZ_18889 [Jatropha curcas] |
| 10 |
Hb_006326_020 |
0.2943011914 |
- |
- |
- |
| 11 |
Hb_060236_010 |
0.3014665441 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 12 |
Hb_004241_050 |
0.3080714375 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 13 |
Hb_007007_020 |
0.3089235686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_041063_010 |
0.3101321936 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_001115_010 |
0.312210034 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |
| 16 |
Hb_001329_030 |
0.31336155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000046_250 |
0.3147100605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_082733_010 |
0.314808484 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 19 |
Hb_137350_010 |
0.3172874118 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 20 |
Hb_003360_050 |
0.3188300373 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |