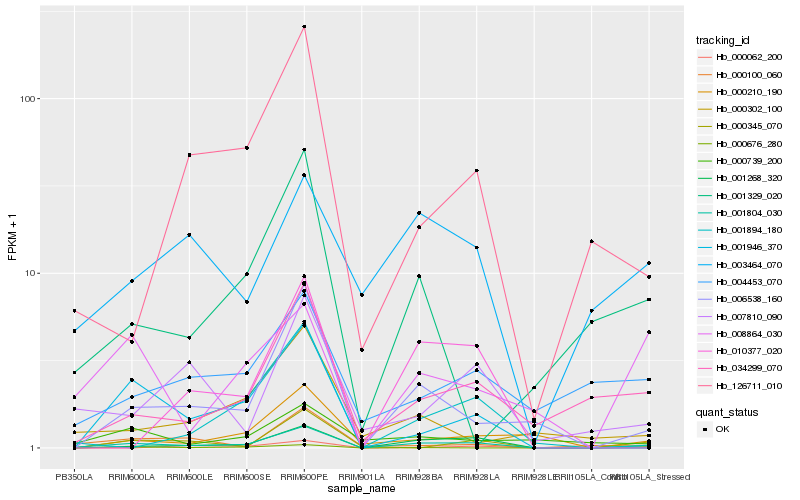
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001804_030 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_006538_160 |
0.2361685308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001268_320 |
0.2531093766 |
- |
- |
PREDICTED: uncharacterized protein LOC104887816 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_034299_070 |
0.2562626337 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 5 |
Hb_001946_370 |
0.2660670571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000210_190 |
0.2840158416 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 7 |
Hb_126711_010 |
0.2879210245 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 8 |
Hb_001894_180 |
0.2946785811 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 9 |
Hb_000345_070 |
0.2969766809 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 10 |
Hb_000676_280 |
0.2986577114 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_004453_070 |
0.2995335685 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_007810_090 |
0.3011627604 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 13 |
Hb_001329_020 |
0.3074487634 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 14 |
Hb_008864_030 |
0.3093379037 |
- |
- |
- |
| 15 |
Hb_000302_100 |
0.3097114516 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 16 |
Hb_003464_070 |
0.310581561 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 17 |
Hb_000100_060 |
0.3118739871 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 18 |
Hb_000062_200 |
0.3128521663 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 19 |
Hb_010377_020 |
0.3143767521 |
- |
- |
- |
| 20 |
Hb_000739_200 |
0.3154459542 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |