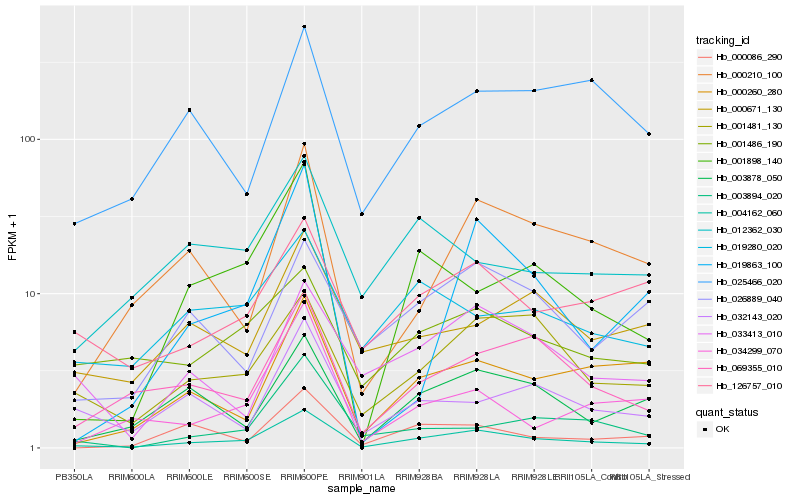
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_060 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_069355_010 |
0.184122838 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 3 |
Hb_001481_130 |
0.1842282609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001898_140 |
0.1871809362 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 5 |
Hb_003878_050 |
0.1930135647 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 6 |
Hb_126757_010 |
0.2003715794 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 7 |
Hb_000210_100 |
0.2017809128 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 8 |
Hb_000260_280 |
0.2018871469 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 9 |
Hb_000086_290 |
0.2039779433 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 10 |
Hb_003894_020 |
0.2042013397 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 11 |
Hb_032143_020 |
0.2077899824 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 12 |
Hb_001486_190 |
0.2081565207 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 13 |
Hb_034299_070 |
0.2114151842 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 14 |
Hb_033413_010 |
0.2126655472 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_019280_020 |
0.2166520423 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 16 |
Hb_025466_020 |
0.219293881 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 17 |
Hb_000671_130 |
0.2208388147 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_026889_040 |
0.2221476183 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 19 |
Hb_012362_030 |
0.2223240566 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 20 |
Hb_019863_100 |
0.2224195779 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |