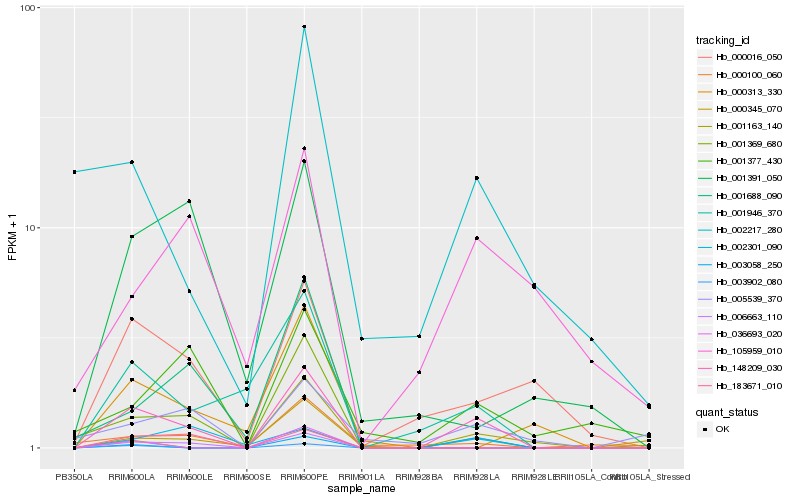
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148209_030 |
0.0 |
- |
- |
hypothetical protein VITISV_017120 [Vitis vinifera] |
| 2 |
Hb_000345_070 |
0.2734397184 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 3 |
Hb_001946_370 |
0.2859767978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006663_110 |
0.292206059 |
- |
- |
PREDICTED: U-box domain-containing protein 34-like [Camelina sativa] |
| 5 |
Hb_000016_050 |
0.3146029515 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 6 |
Hb_003058_250 |
0.3228422893 |
- |
- |
hypothetical protein RCOM_0006440 [Ricinus communis] |
| 7 |
Hb_002301_090 |
0.3330528308 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 8 |
Hb_001369_680 |
0.336876516 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 9 |
Hb_000100_060 |
0.3380019291 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 10 |
Hb_002217_280 |
0.3506195244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_183671_010 |
0.3509184475 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_005539_370 |
0.3536590004 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_000313_330 |
0.3557401551 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 14 |
Hb_001163_140 |
0.3604314753 |
- |
- |
- |
| 15 |
Hb_001377_430 |
0.3625727974 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 16 |
Hb_001391_050 |
0.3679010337 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 17 |
Hb_105959_010 |
0.3686677708 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 18 |
Hb_036693_020 |
0.3706172304 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 19 |
Hb_003902_080 |
0.3750822503 |
- |
- |
PREDICTED: senescence-specific cysteine protease SAG39-like [Populus euphratica] |
| 20 |
Hb_001688_090 |
0.3750934635 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |