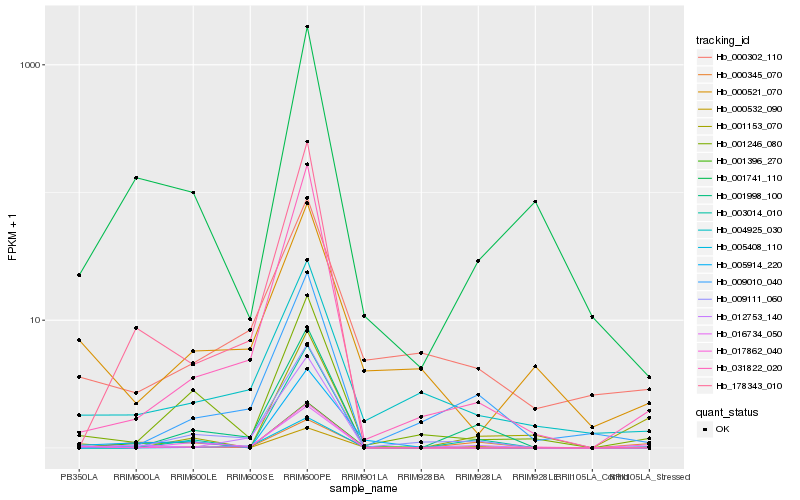
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000532_090 |
0.0 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 2 |
Hb_005914_220 |
0.2175885999 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000302_110 |
0.218587136 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004925_030 |
0.2204943137 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 5 |
Hb_017862_040 |
0.224475668 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 6 |
Hb_001741_110 |
0.2259105534 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 7 |
Hb_009111_060 |
0.2269352951 |
- |
- |
PREDICTED: sodium transporter HKT1-like [Jatropha curcas] |
| 8 |
Hb_031822_020 |
0.2276054335 |
- |
- |
PREDICTED: uncharacterized protein LOC104613080 [Nelumbo nucifera] |
| 9 |
Hb_001246_080 |
0.232062633 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 10 |
Hb_001998_100 |
0.2341582419 |
- |
- |
- |
| 11 |
Hb_001396_270 |
0.2342194247 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 12 |
Hb_016734_050 |
0.2447029647 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 13 |
Hb_005408_110 |
0.2513063987 |
- |
- |
purine permease, putative [Ricinus communis] |
| 14 |
Hb_003014_010 |
0.2519115929 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0017s11800g [Populus trichocarpa] |
| 15 |
Hb_012753_140 |
0.2523169121 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 16 |
Hb_009010_040 |
0.2530566238 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 17 |
Hb_000345_070 |
0.2531339226 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 18 |
Hb_000521_070 |
0.2568770118 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 19 |
Hb_001153_070 |
0.2569897649 |
- |
- |
- |
| 20 |
Hb_178343_010 |
0.2574906879 |
- |
- |
PREDICTED: two-component response regulator ARR17-like [Jatropha curcas] |