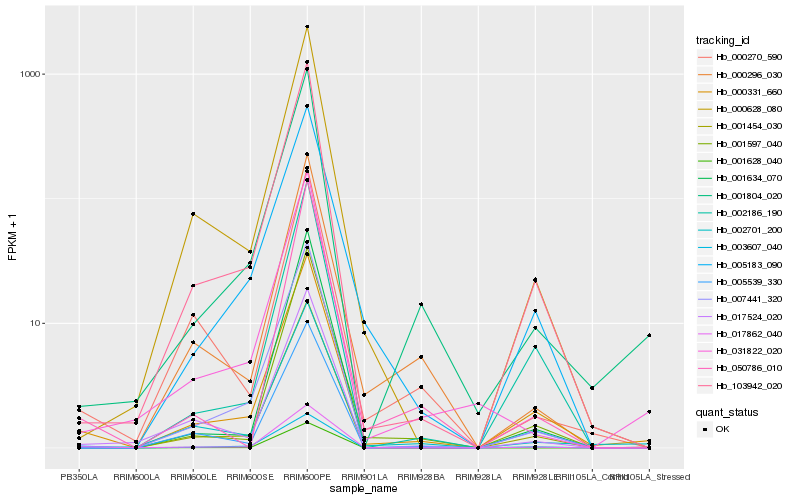
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017862_040 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 2 |
Hb_000331_660 |
0.0897588902 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 3 |
Hb_002701_200 |
0.125933456 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 4 |
Hb_031822_020 |
0.1261040293 |
- |
- |
PREDICTED: uncharacterized protein LOC104613080 [Nelumbo nucifera] |
| 5 |
Hb_001804_020 |
0.1294474094 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 6 |
Hb_103942_020 |
0.1355029506 |
- |
- |
unknown [Zea mays] |
| 7 |
Hb_017524_020 |
0.1384319644 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 8 |
Hb_000628_080 |
0.1425208403 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 9 |
Hb_001597_040 |
0.1428503663 |
- |
- |
- |
| 10 |
Hb_050786_010 |
0.146118276 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |
| 11 |
Hb_001454_030 |
0.1464228608 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 12 |
Hb_007441_320 |
0.1498469325 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 13 |
Hb_005539_330 |
0.150948021 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 14 |
Hb_001634_070 |
0.1516894624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001628_040 |
0.1520909397 |
- |
- |
PREDICTED: beta-galactosidase 13-like [Populus euphratica] |
| 16 |
Hb_005183_090 |
0.1522430265 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 17 |
Hb_000296_030 |
0.158934993 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 18 |
Hb_003607_040 |
0.1596099969 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000270_590 |
0.1599663119 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 20 |
Hb_002186_190 |
0.1611246246 |
- |
- |
- |