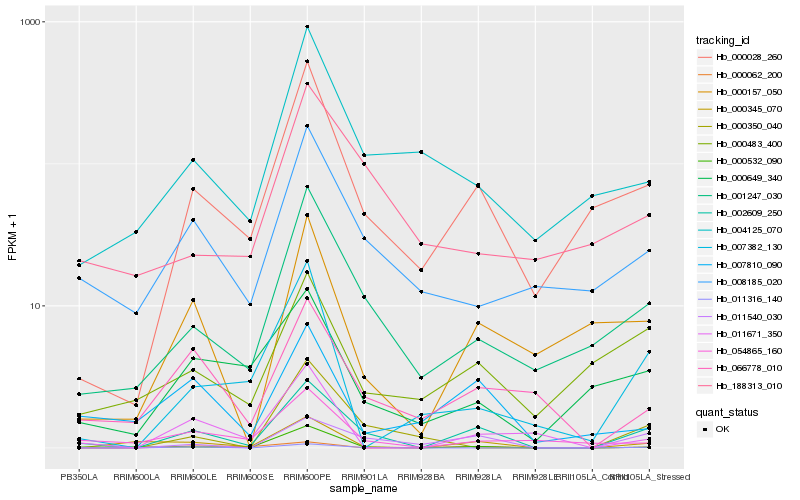
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_250 |
0.0 |
- |
- |
PREDICTED: starch-binding domain-containing protein 1-like [Jatropha curcas] |
| 2 |
Hb_011540_030 |
0.2476703164 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 3 |
Hb_000028_260 |
0.2491305272 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 4 |
Hb_001247_030 |
0.2593477391 |
- |
- |
- |
| 5 |
Hb_000345_070 |
0.2831699732 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 6 |
Hb_000532_090 |
0.2923205469 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 7 |
Hb_011316_140 |
0.2950673053 |
- |
- |
PREDICTED: uncharacterized protein LOC105129591 [Populus euphratica] |
| 8 |
Hb_066778_010 |
0.2994062787 |
- |
- |
- |
| 9 |
Hb_000157_050 |
0.3025091554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008185_020 |
0.3068796914 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 11 |
Hb_007810_090 |
0.3141517696 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 12 |
Hb_054865_160 |
0.3180898239 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 13 |
Hb_004125_070 |
0.3192249267 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 14 |
Hb_000483_400 |
0.3199451717 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_011671_350 |
0.3200167649 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 16 |
Hb_000649_340 |
0.3218253739 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_007382_130 |
0.3273810985 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 18 |
Hb_000350_040 |
0.3280276867 |
- |
- |
- |
| 19 |
Hb_000062_200 |
0.3305844454 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_188313_010 |
0.3308902373 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |