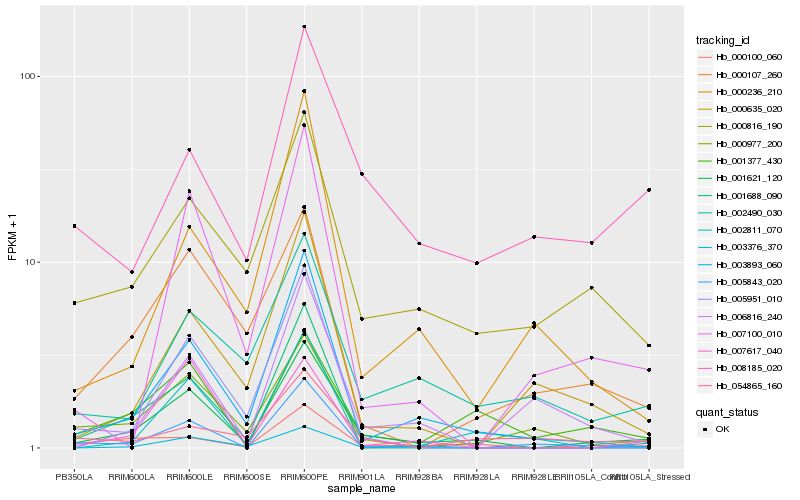
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_120 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_000100_060 |
0.1836718338 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 3 |
Hb_003893_060 |
0.1944943821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001377_430 |
0.2063650343 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 5 |
Hb_003376_370 |
0.2156792201 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000977_200 |
0.2164931985 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000816_190 |
0.2213842214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002490_030 |
0.2336280031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007100_010 |
0.2376215519 |
- |
- |
hypothetical protein ARALYDRAFT_479822 [Arabidopsis lyrata subsp. lyrata] |
| 10 |
Hb_000107_260 |
0.2428390308 |
- |
- |
- |
| 11 |
Hb_002811_070 |
0.243839135 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_005843_020 |
0.2452852281 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_000236_210 |
0.2469713332 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 14 |
Hb_000635_020 |
0.2480610984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001688_090 |
0.2507218982 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 16 |
Hb_007617_040 |
0.2507525408 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 17 |
Hb_005951_010 |
0.2513393137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_054865_160 |
0.2523820501 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 19 |
Hb_008185_020 |
0.2523830432 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 20 |
Hb_006816_240 |
0.2531705707 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |