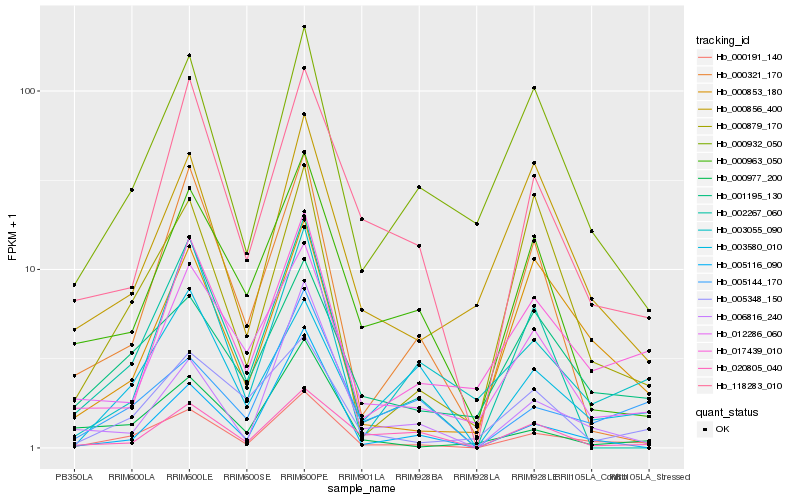
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_140 |
0.0 |
- |
- |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_118283_010 |
0.1788957185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001195_130 |
0.1811231031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005144_170 |
0.1814316856 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_017439_010 |
0.1867405596 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 6 |
Hb_012286_060 |
0.1932088028 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000932_050 |
0.1965942452 |
- |
- |
annexin [Manihot esculenta] |
| 8 |
Hb_000853_180 |
0.1966963026 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_003055_090 |
0.1967907573 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 10 |
Hb_000977_200 |
0.2000601073 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002267_060 |
0.20071519 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 12 |
Hb_005348_150 |
0.2031510871 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 13 |
Hb_003580_010 |
0.204038226 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 14 |
Hb_000963_050 |
0.2104986862 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 15 |
Hb_000856_400 |
0.2115498353 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 16 |
Hb_005116_090 |
0.2125021312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000879_170 |
0.212937923 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_006816_240 |
0.2129546763 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 19 |
Hb_020805_040 |
0.213222506 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1A [Jatropha curcas] |
| 20 |
Hb_000321_170 |
0.2153940426 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |