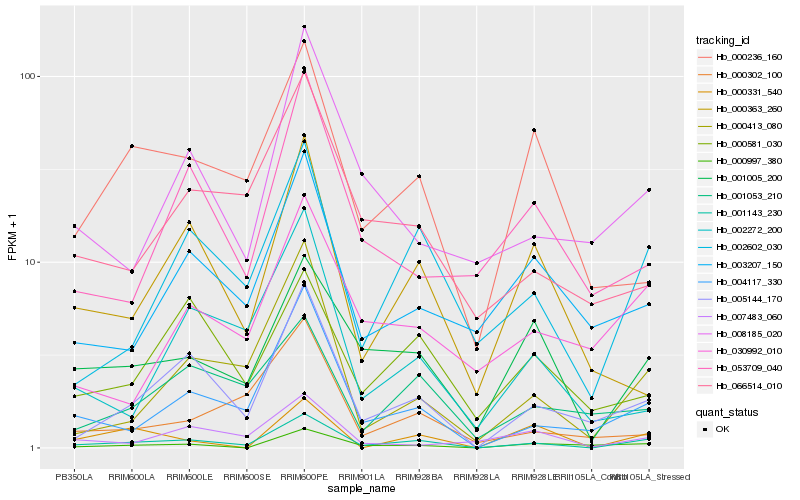
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_230 |
0.0 |
- |
- |
beta-galactosidase family protein [Populus trichocarpa] |
| 2 |
Hb_005144_170 |
0.1829609066 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_004117_330 |
0.1851131779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002602_030 |
0.199947716 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001005_200 |
0.208778212 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000413_080 |
0.2096933299 |
- |
- |
- |
| 7 |
Hb_000997_380 |
0.2411131083 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 8 |
Hb_000236_160 |
0.2455332784 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 9 |
Hb_001053_210 |
0.2507307188 |
- |
- |
- |
| 10 |
Hb_000363_260 |
0.251524787 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 11 |
Hb_066514_010 |
0.2515404725 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 12 |
Hb_003207_150 |
0.2522011639 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 13 |
Hb_008185_020 |
0.2522573867 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 14 |
Hb_030992_010 |
0.2563109703 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_002272_200 |
0.2579616545 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000581_030 |
0.261026358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007483_060 |
0.2620455343 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 18 |
Hb_000302_100 |
0.2625040469 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 19 |
Hb_053709_040 |
0.2630669775 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 20 |
Hb_000331_540 |
0.2637630889 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |