| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
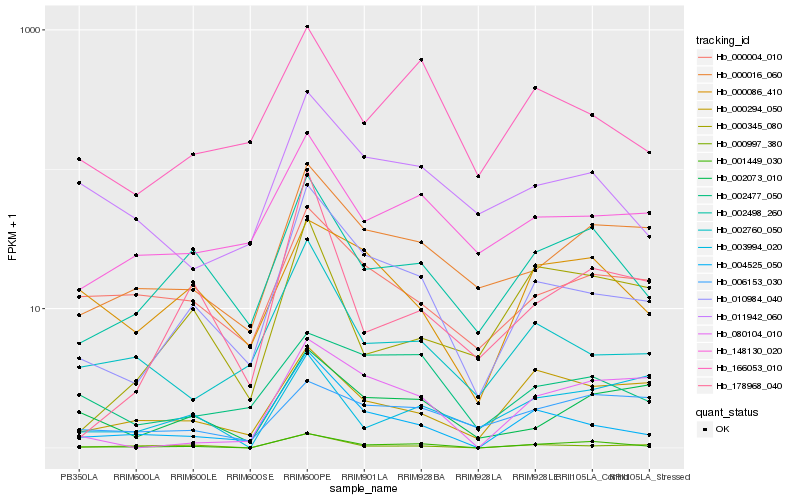
Hb_001449_030 |
0.0 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002498_260 |
0.198225298 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 3 |
Hb_010984_040 |
0.2002535908 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 4 |
Hb_004525_050 |
0.2085364126 |
- |
- |
similar to Drosophila melanogaster CG10960, partial [Drosophila yakuba] |
| 5 |
Hb_000997_380 |
0.2173398474 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 6 |
Hb_000016_060 |
0.2327180504 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 7 |
Hb_000294_050 |
0.2410924184 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 8 |
Hb_002073_010 |
0.2469320844 |
- |
- |
- |
| 9 |
Hb_080104_010 |
0.2497785126 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000086_410 |
0.2526787336 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 11 |
Hb_166053_010 |
0.2565343988 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 12 |
Hb_011942_060 |
0.2588409236 |
- |
- |
hypothetical protein CISIN_1g028965mg [Citrus sinensis] |
| 13 |
Hb_002760_050 |
0.2603534007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003994_020 |
0.2611927007 |
- |
- |
- |
| 15 |
Hb_000004_010 |
0.2626282508 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 16 |
Hb_000345_080 |
0.2633934556 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 17 |
Hb_148130_020 |
0.2650394497 |
- |
- |
- |
| 18 |
Hb_178968_040 |
0.2653923653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002477_050 |
0.2677625869 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_006153_030 |
0.2681926232 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |