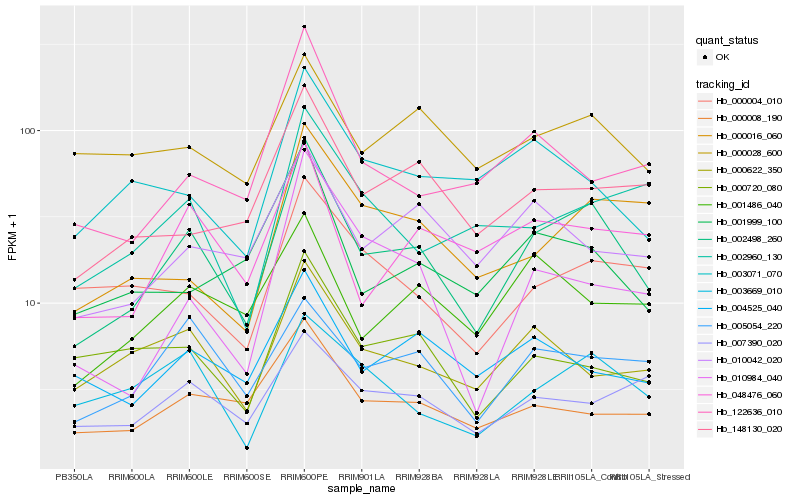
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_260 |
0.0 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 2 |
Hb_048476_060 |
0.1700501908 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_005054_220 |
0.1745701905 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 4 |
Hb_001999_100 |
0.1752546684 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 5 |
Hb_000008_190 |
0.1753572098 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_148130_020 |
0.175613571 |
- |
- |
- |
| 7 |
Hb_003669_010 |
0.1791538585 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 8 |
Hb_010042_020 |
0.1821966862 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 9 |
Hb_000622_350 |
0.1827872291 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 10 |
Hb_001486_040 |
0.1829234379 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000016_060 |
0.1843887686 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_004525_040 |
0.1862419114 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 13 |
Hb_010984_040 |
0.1869959517 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 14 |
Hb_003071_070 |
0.1873226759 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 15 |
Hb_000028_600 |
0.1877002294 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 16 |
Hb_000004_010 |
0.190735895 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 17 |
Hb_122636_010 |
0.191284344 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 18 |
Hb_000720_080 |
0.1921265078 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 19 |
Hb_002960_130 |
0.1922426663 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 20 |
Hb_007390_020 |
0.1938016614 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |