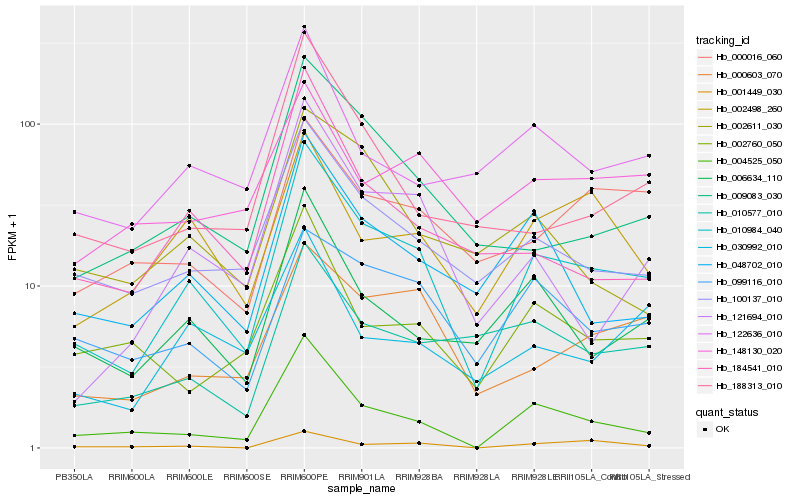
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010984_040 |
0.0 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 2 |
Hb_004525_050 |
0.151252423 |
- |
- |
similar to Drosophila melanogaster CG10960, partial [Drosophila yakuba] |
| 3 |
Hb_100137_010 |
0.1565909018 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 4 |
Hb_006634_110 |
0.1719893774 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 5 |
Hb_009083_030 |
0.1727383349 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 6 |
Hb_048702_010 |
0.1735545724 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_121694_010 |
0.1742084219 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 8 |
Hb_000603_070 |
0.1770075184 |
- |
- |
- |
| 9 |
Hb_099116_010 |
0.1858541698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002498_260 |
0.1869959517 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 11 |
Hb_122636_010 |
0.1897097739 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 12 |
Hb_010577_010 |
0.1903578465 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 13 |
Hb_030992_010 |
0.1912517076 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 14 |
Hb_000016_060 |
0.1923372822 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 15 |
Hb_188313_010 |
0.1927794683 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 16 |
Hb_002760_050 |
0.1947319887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_184541_010 |
0.1988291232 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 18 |
Hb_001449_030 |
0.2002535908 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_148130_020 |
0.2022969583 |
- |
- |
- |
| 20 |
Hb_002611_030 |
0.205576987 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |