| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
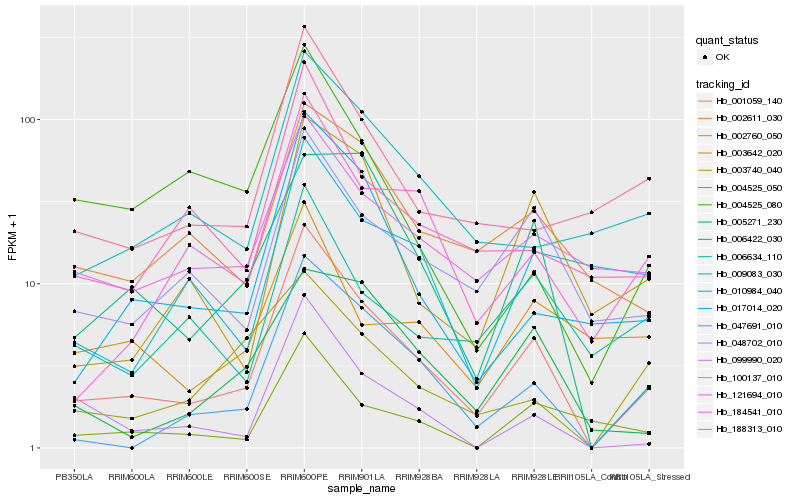
Hb_001059_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 2 |
Hb_047691_010 |
0.1691903622 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 3 |
Hb_048702_010 |
0.1837252501 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_017014_020 |
0.1875986478 |
- |
- |
- |
| 5 |
Hb_121694_010 |
0.1895551514 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 6 |
Hb_099990_020 |
0.1941672552 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 7 |
Hb_184541_010 |
0.1999170136 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 8 |
Hb_100137_010 |
0.2009082997 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 9 |
Hb_188313_010 |
0.2014469772 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 10 |
Hb_009083_030 |
0.2015379601 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 11 |
Hb_002611_030 |
0.2069716623 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 12 |
Hb_003642_020 |
0.2103398588 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 13 |
Hb_004525_050 |
0.2127624298 |
- |
- |
similar to Drosophila melanogaster CG10960, partial [Drosophila yakuba] |
| 14 |
Hb_006634_110 |
0.2175694694 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 15 |
Hb_006422_030 |
0.2270713532 |
- |
- |
- |
| 16 |
Hb_005271_230 |
0.2276027728 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 17 |
Hb_003740_040 |
0.2297017364 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 18 |
Hb_004525_080 |
0.2323103198 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 19 |
Hb_002760_050 |
0.2331624887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010984_040 |
0.2348573595 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |