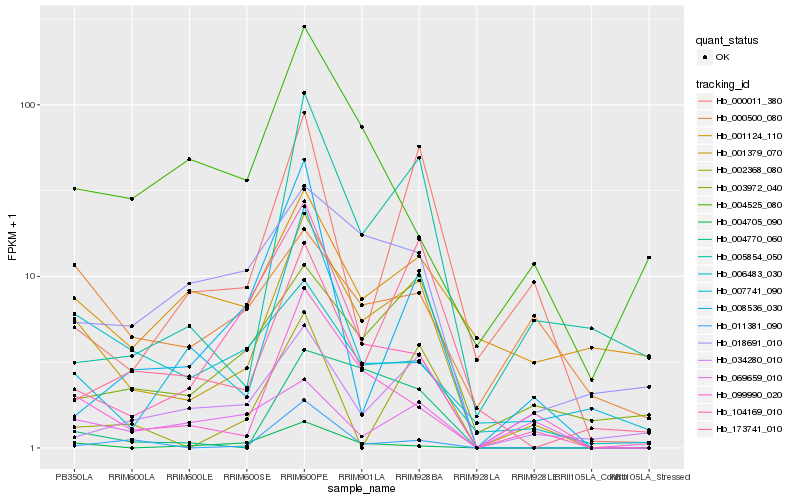
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001379_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_16377 [Jatropha curcas] |
| 2 |
Hb_099990_020 |
0.2328627634 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 3 |
Hb_003972_040 |
0.2556911624 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 4 |
Hb_018691_010 |
0.263216854 |
- |
- |
- |
| 5 |
Hb_005854_050 |
0.2635655568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_104169_010 |
0.2687238229 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_034280_010 |
0.2700231625 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 8 |
Hb_004705_090 |
0.2736605746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000011_380 |
0.2797038183 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 10 |
Hb_004770_060 |
0.2824401244 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 11 |
Hb_006483_030 |
0.2863826229 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 12 |
Hb_001124_110 |
0.2930779071 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_007741_090 |
0.2933117128 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 14 |
Hb_173741_010 |
0.2940283105 |
- |
- |
PREDICTED: caffeoyl-CoA O-methyltransferase isoform X3 [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_008536_030 |
0.2978211793 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 16 |
Hb_069659_010 |
0.2997238796 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_000500_080 |
0.3003506764 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 18 |
Hb_011381_090 |
0.3004708553 |
- |
- |
Protein ABC1, mitochondrial precursor, putative [Ricinus communis] |
| 19 |
Hb_004525_080 |
0.3032887176 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 20 |
Hb_002368_080 |
0.304220195 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X2 [Jatropha curcas] |