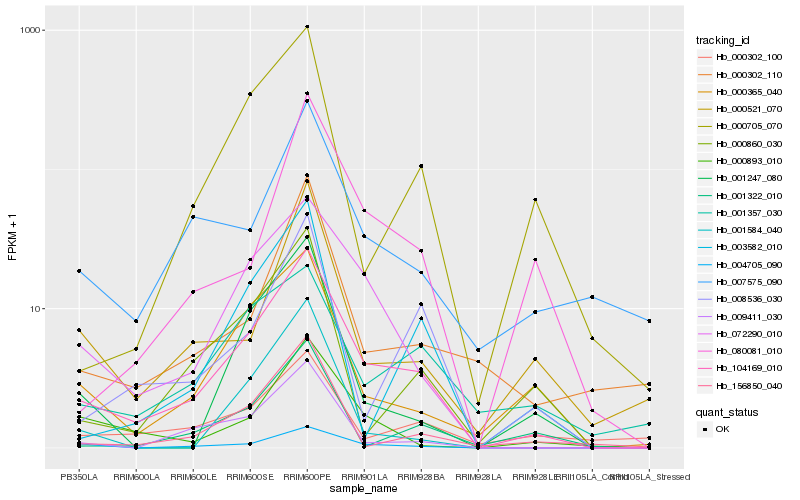
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104169_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_072290_010 |
0.1599204927 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 3 |
Hb_004705_090 |
0.1726254435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000365_040 |
0.1952441256 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 5 |
Hb_009411_030 |
0.1988221319 |
- |
- |
PREDICTED: bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3-like [Malus domestica] |
| 6 |
Hb_000860_030 |
0.2124012241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008536_030 |
0.2179693859 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 8 |
Hb_007575_090 |
0.2187118269 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 9 |
Hb_000893_010 |
0.2187904638 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 10 |
Hb_003582_010 |
0.219457535 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 11 |
Hb_001357_030 |
0.219960557 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 12 |
Hb_000705_070 |
0.2213342007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_156850_040 |
0.2213714962 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 14 |
Hb_001247_080 |
0.223202753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000302_110 |
0.2260068474 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_080081_010 |
0.2261053829 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000521_070 |
0.2308707122 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 18 |
Hb_000302_100 |
0.2320399441 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 19 |
Hb_001322_010 |
0.2340374992 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 20 |
Hb_001584_040 |
0.2367602761 |
- |
- |
PREDICTED: uncharacterized protein LOC105125166 [Populus euphratica] |