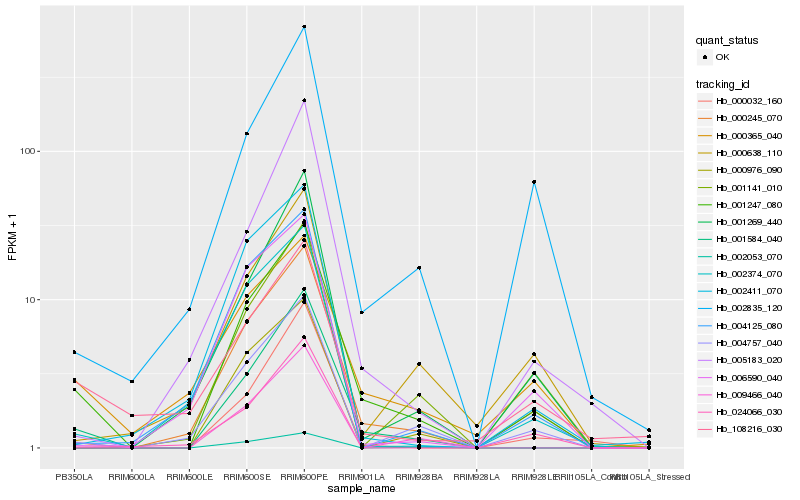
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001584_040 |
0.1093424669 |
- |
- |
PREDICTED: uncharacterized protein LOC105125166 [Populus euphratica] |
| 3 |
Hb_000245_070 |
0.1292471432 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 4 |
Hb_004757_040 |
0.1697299291 |
- |
- |
- |
| 5 |
Hb_000365_040 |
0.1757673397 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 6 |
Hb_002835_120 |
0.1837940143 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 7 |
Hb_108216_030 |
0.1873621623 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001269_440 |
0.1964862087 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 9 |
Hb_002374_070 |
0.19911012 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000638_110 |
0.199730579 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 11 |
Hb_001141_010 |
0.1999253452 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 12 |
Hb_005183_020 |
0.2017072672 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |
| 13 |
Hb_004125_080 |
0.2042571065 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 14 |
Hb_009466_040 |
0.2054348565 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 15 |
Hb_000976_090 |
0.2057969828 |
- |
- |
- |
| 16 |
Hb_006590_040 |
0.2071554666 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 17 |
Hb_000032_160 |
0.2072965497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_024066_030 |
0.2084505743 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 19 |
Hb_002411_070 |
0.2086743875 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 20 |
Hb_002053_070 |
0.2097550395 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |