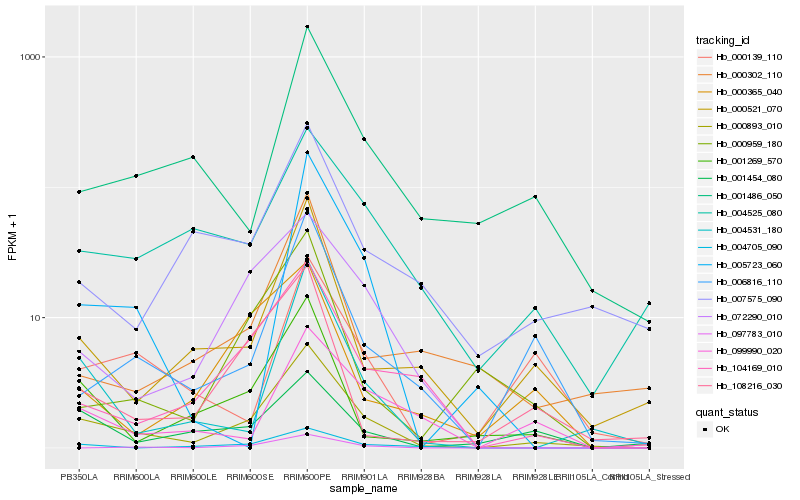
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000893_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 2 |
Hb_004705_090 |
0.214066491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004531_180 |
0.216376551 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_104169_010 |
0.2187904638 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_072290_010 |
0.2209208805 |
- |
- |
PREDICTED: GRF1-interacting factor 1 [Gossypium raimondii] |
| 6 |
Hb_006816_110 |
0.2216823466 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 7 |
Hb_000521_070 |
0.2232926427 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 8 |
Hb_001269_570 |
0.2293105066 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 9 |
Hb_004525_080 |
0.2363812189 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 10 |
Hb_108216_030 |
0.237690652 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_097783_010 |
0.2381427084 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001486_050 |
0.2405280861 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_099990_020 |
0.2452107989 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 14 |
Hb_007575_090 |
0.2525443755 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 15 |
Hb_000139_110 |
0.2533930012 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 16 |
Hb_001454_080 |
0.2554237835 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_005723_060 |
0.2579863011 |
- |
- |
- |
| 18 |
Hb_000959_180 |
0.2596742565 |
- |
- |
hypothetical protein JCGZ_09448 [Jatropha curcas] |
| 19 |
Hb_000365_040 |
0.2615234305 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 20 |
Hb_000302_110 |
0.2620765033 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |