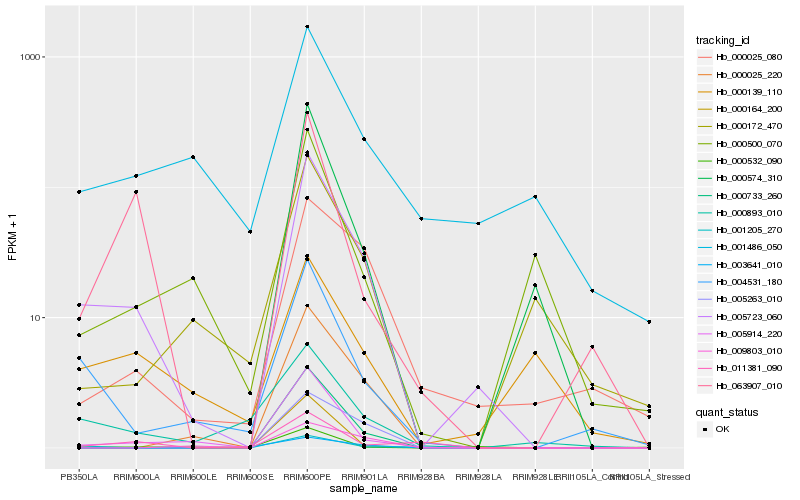
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005723_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_005914_220 |
0.2050340975 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004531_180 |
0.2155748281 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_000733_260 |
0.2414753745 |
- |
- |
hypothetical protein JCGZ_26934 [Jatropha curcas] |
| 5 |
Hb_000025_220 |
0.2430724964 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 6 |
Hb_000025_080 |
0.2507687926 |
- |
- |
- |
| 7 |
Hb_001205_270 |
0.2559914865 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 8 |
Hb_000893_010 |
0.2579863011 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 9 |
Hb_000532_090 |
0.259439034 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 10 |
Hb_005263_010 |
0.2639962304 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 11 |
Hb_063907_010 |
0.2689898133 |
- |
- |
- |
| 12 |
Hb_001486_050 |
0.2714920492 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_000574_310 |
0.2824146454 |
- |
- |
hypothetical protein CISIN_1g027539mg [Citrus sinensis] |
| 14 |
Hb_000172_470 |
0.2846439342 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 15 |
Hb_011381_090 |
0.2873149847 |
- |
- |
Protein ABC1, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_000139_110 |
0.2883193788 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 17 |
Hb_000164_200 |
0.2903248274 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 18 |
Hb_000500_070 |
0.2920230378 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 19 |
Hb_009803_010 |
0.2940955809 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 20 |
Hb_003641_010 |
0.2943880181 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |