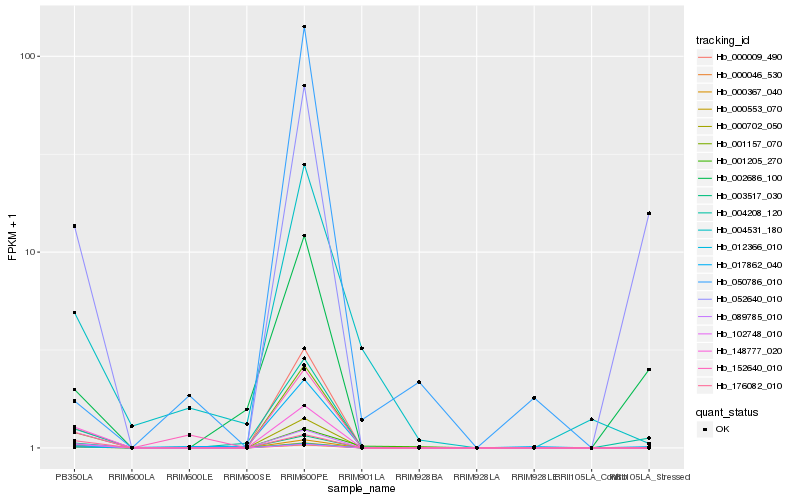
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012366_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 2 |
Hb_089785_010 |
0.0082154575 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 3 |
Hb_003517_030 |
0.0915216397 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 4 |
Hb_001157_070 |
0.0934385008 |
- |
- |
- |
| 5 |
Hb_000009_490 |
0.09862511 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 6 |
Hb_148777_020 |
0.11491369 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 7 |
Hb_176082_010 |
0.1523443276 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 8 |
Hb_000702_050 |
0.2004060957 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000367_040 |
0.2151360572 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_004208_120 |
0.2154982919 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 11 |
Hb_152640_010 |
0.2265666308 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 12 |
Hb_001205_270 |
0.2470732132 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 13 |
Hb_004531_180 |
0.2530160288 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_102748_010 |
0.2744755038 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 15 |
Hb_052640_010 |
0.2856895542 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000553_070 |
0.2882087293 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X2 [Jatropha curcas] |
| 17 |
Hb_017862_040 |
0.29932383 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 18 |
Hb_000046_530 |
0.3049023943 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 19 |
Hb_002686_100 |
0.3052246981 |
- |
- |
- |
| 20 |
Hb_050786_010 |
0.3069643902 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |