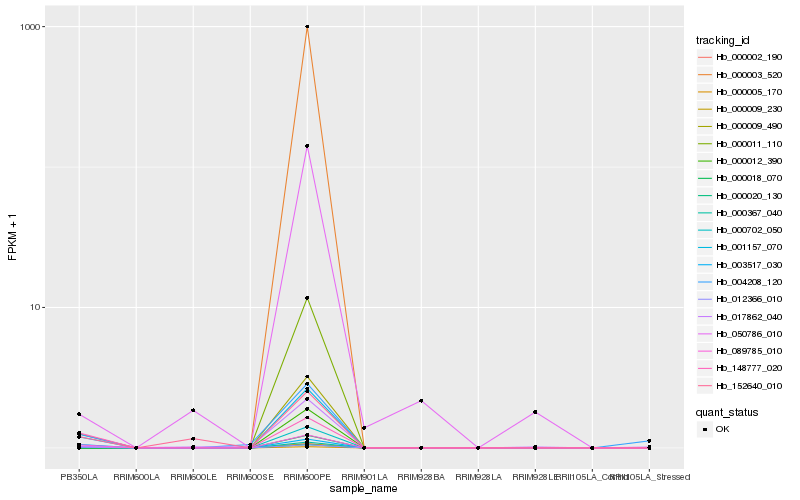
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000009_490 |
0.0052437499 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 3 |
Hb_089785_010 |
0.0852868681 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 4 |
Hb_012366_010 |
0.0934385008 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 5 |
Hb_000702_050 |
0.1100617216 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003517_030 |
0.1836991998 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 7 |
Hb_000367_040 |
0.1837397097 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_004208_120 |
0.2016944989 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 9 |
Hb_148777_020 |
0.2066457818 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 10 |
Hb_152640_010 |
0.2129684633 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 11 |
Hb_050786_010 |
0.2317965425 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |
| 12 |
Hb_017862_040 |
0.2358267561 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 13 |
Hb_000002_190 |
0.2366548683 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 14 |
Hb_000003_520 |
0.2366548683 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 15 |
Hb_000005_170 |
0.2366548683 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR [Jatropha curcas] |
| 16 |
Hb_000009_230 |
0.2366548683 |
- |
- |
hypothetical protein POPTR_0007s12290g [Populus trichocarpa] |
| 17 |
Hb_000011_110 |
0.2366548683 |
- |
- |
PREDICTED: uncharacterized protein LOC105631299 [Jatropha curcas] |
| 18 |
Hb_000012_390 |
0.2366548683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000018_070 |
0.2366548683 |
- |
- |
PREDICTED: amino acid permease 8-like [Jatropha curcas] |
| 20 |
Hb_000020_130 |
0.2366548683 |
- |
- |
conidial pigment biosynthesis oxidase Arb2/brown2 [Aspergillus clavatus NRRL 1] |