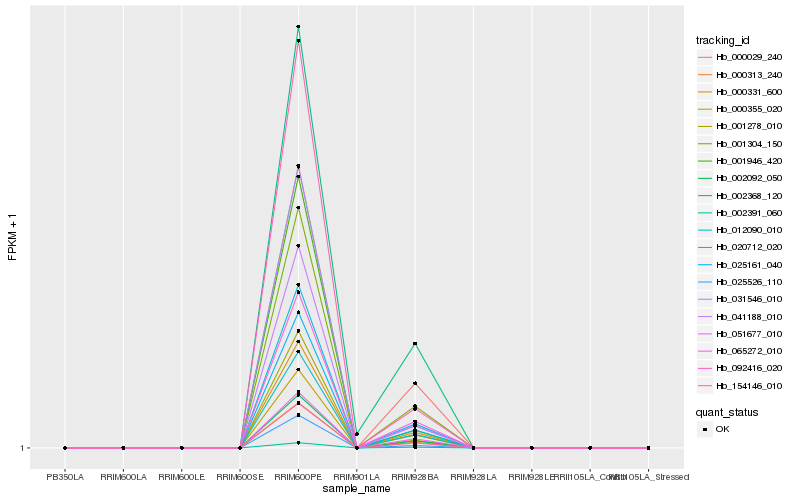
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020712_020 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Elaeis guineensis] |
| 2 |
Hb_025526_110 |
0.0088358479 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 3 |
Hb_000355_020 |
0.0092417453 |
- |
- |
- |
| 4 |
Hb_001278_010 |
0.0116156441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_041188_010 |
0.0121724814 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Nicotiana tomentosiformis] |
| 6 |
Hb_154146_010 |
0.0320957286 |
- |
- |
hypothetical protein JCGZ_00346 [Jatropha curcas] |
| 7 |
Hb_001304_150 |
0.0349366143 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_000313_240 |
0.0469074334 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 9 |
Hb_012090_010 |
0.0474358115 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 10 |
Hb_000331_600 |
0.0502510457 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 11 |
Hb_002092_050 |
0.0529428593 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 12 |
Hb_065272_010 |
0.0571812735 |
- |
- |
hypothetical protein JCGZ_06349 [Jatropha curcas] |
| 13 |
Hb_031546_010 |
0.0605837196 |
- |
- |
hypothetical protein JCGZ_21533 [Jatropha curcas] |
| 14 |
Hb_000029_240 |
0.0623057901 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Elaeis guineensis] |
| 15 |
Hb_025161_040 |
0.0688844181 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_092416_020 |
0.0717379256 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001946_420 |
0.0752344166 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_002368_120 |
0.0768005449 |
- |
- |
hypothetical protein VITISV_033181 [Vitis vinifera] |
| 19 |
Hb_002391_060 |
0.0802545366 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 20 |
Hb_051677_010 |
0.0810448506 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |